Insulin-induced formation of macromolecular complexes involved in activation of cyclic nucleotide phosphodiesterase 3B (PDE3B) and its interaction with PKB
- PMID: 17324123
- PMCID: PMC1868803
- DOI: 10.1042/BJ20060960
Insulin-induced formation of macromolecular complexes involved in activation of cyclic nucleotide phosphodiesterase 3B (PDE3B) and its interaction with PKB
Abstract
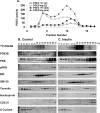
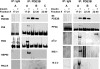
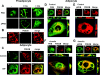
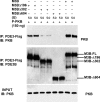
Fractionation of 3T3-L1 adipocyte membranes revealed that PDE3B (phosphodiesterase 3B) was associated with PM (plasma membrane) and ER (endoplasmic reticulum)/Golgi fractions, that insulin-induced phosphorylation/activation of PDE3B was greater in internal membranes than PM fractions, and that there was no significant translocation of PDE3B between membrane fractions. Insulin also induced formation of large macromolecular complexes, separated during gel filtration (Superose 6 columns) of solubilized membranes, which apparently contain phosphorylated/activated PDE3B and signalling molecules potentially involved in its activation by insulin, e.g. IRS-1 (insulin receptor substrate-1), IRS-2, PI3K p85 [p85-subunit of PI3K (phosphoinositide 3-kinase)], PKB (protein kinase B), HSP-90 (heat-shock protein 90) and 14-3-3. Expression of full-length recombinant FLAG-tagged murine (M) PDE3B and M3BDelta604 (MPDE3B lacking N-terminal 604 amino acids) indicated that the N-terminal region of MPDE3B was necessary for insulin-induced activation and recruitment of PDE3B. siRNA (small interfering RNA) knock-down of PDE3B indicated that PDE3B was not required for formation of insulin-induced complexes. Wortmannin inhibited insulin-induced assembly of macromolecular complexes, as well as phosphorylation/activation of PKB and PDE3B, and their co-immunoprecipitation. Another PI3K inhibitor, LY294002, and the tyrosine kinase inhibitor, Genistein, also inhibited insulin-induced activation of PDE3B and its co-immunoprecipitation with PKB. Confocal microscopy indicated co-localization of PDE3B and PKB. Recombinant MPDE3B co-immunoprecipitated, and co-eluted during Superose 12 chromatography, to a greater extent with recombinant pPKB (phosphorylated/activated PKB) than dephospho-PKB or p-DeltaPKB [pPKB lacking its PH domain (pleckstrin homology domain)]. Truncated recombinant MPDE3B proteins and pPKB did not efficiently co-immunoprecipitate, suggesting that structural determinants for their interaction reside in, or are regulated by, the N-terminal portion of MPDE3B. Recruitment of PDE3B in macromolecular complexes may be critical for regulation of specific cAMP pools and signalling pathways by insulin, e.g. lipolysis.
Figures






References
-
- Francis S. H., Turko I. V., Corbin J. D. Cyclic nucleotide phosphodiesterases: relating structure and function. Prog. Nucleic Acid. Res. Mol. Biol. 2001;65:1–52. - PubMed
-
- Shakur Y., Holst L. S., Landstrom T. R., Movsesian M., Degerman E., Manganiello V. Regulation and function of the cyclic nucleotide phosphodiesterase (PDE3) gene family. Prog. Nucleic Acid Res. Mol. Biol. 2001;66:241–277. - PubMed
-
- Walz H. A., Harndahl L., Wierup N., Zmuda-Trzebiatowska E., Svennelid F., Manganiello V. C., Ploug T., Sundler F., Degerman E., Ahren B., et al. Early and rapid development of insulin resistance, islet dysfunction and glucose intolerance after high-fat feeding in mice overexpressing phosphodiesterase 3B. J. Endocrinol. 2006;189:629–641. - PubMed
-
- Houslay M. D., Milligan G. Tailoring cAMP-signalling responses through isoform multiplicity. Trends Biochem. Sci. 1997;22:217–224. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

