HOXA10 controls osteoblastogenesis by directly activating bone regulatory and phenotypic genes
- PMID: 17325044
- PMCID: PMC1899966
- DOI: 10.1128/MCB.01544-06
HOXA10 controls osteoblastogenesis by directly activating bone regulatory and phenotypic genes
Abstract
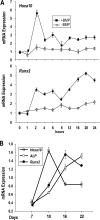
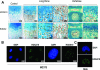
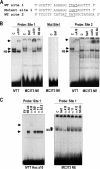
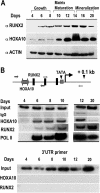
HOXA10 is necessary for embryonic patterning of skeletal elements, but its function in bone formation beyond this early developmental stage is unknown. Here we show that HOXA10 contributes to osteogenic lineage determination through activation of Runx2 and directly regulates osteoblastic phenotypic genes. In response to bone morphogenic protein BMP2, Hoxa10 is rapidly induced and functions to activate the Runx2 transcription factor essential for bone formation. A functional element with the Hox core motif was characterized for the bone-related Runx2 P1 promoter. HOXA10 also activates other osteogenic genes, including the alkaline phosphatase, osteocalcin, and bone sialoprotein genes, and temporally associates with these target gene promoters during stages of osteoblast differentiation prior to the recruitment of RUNX2. Exogenous expression and small interfering RNA knockdown studies establish that HOXA10 mediates chromatin hyperacetylation and trimethyl histone K4 (H3K4) methylation of these genes, correlating to active transcription. HOXA10 therefore contributes to early expression of osteogenic genes through chromatin remodeling. Importantly, HOXA10 can induce osteoblast genes in Runx2 null cells, providing evidence for a direct role in mediating osteoblast differentiation independent of RUNX2. We propose that HOXA10 activates RUNX2 in mesenchymal cells, contributing to the onset of osteogenesis, and that HOXA10 subsequently supports bone formation by direct regulation of osteoblast phenotypic genes.
Figures






References
-
- Bae, J.-S., S. Gutierrez, R. Narla, J. Pratap, R. Devados, J. L. Stein, J. B. Lian, G. S. Stein, and A. Javed. 2007. Reconstitution of Runx2/Cbfa1 null cells identifies Runx2 functional domains required for osteoblast differentiation and responsiveness to osteogenic regulators BMP2, TGFα and 1,25(OH)2D3. J. Cell. Biochem. 100:434-449. - PubMed
-
- Bae, N. S., M. J. Swanson, A. Vassilev, and B. H. Howard. 2004. Human histone deacetylase SIRT2 interacts with the homeobox transcription factor HOXA10. J. Biochem. (Tokyo) 135:695-700. - PubMed
-
- Balint, E., D. Lapointe, H. Drissi, C. van der Meijden, D. W. Young, A. J. van Wijnen, J. L. Stein, G. S. Stein, and J. B. Lian. 2003. Phenotype discovery by gene expression profiling: mapping of biological processes linked to BMP-2-mediated osteoblast differentiation. J. Cell. Biochem. 89:401-426. - PubMed
-
- Bondos, S. 2006. Variations on a theme: Hox and Wnt combinatorial regulation during animal development. Sci. STKE 2006:e38. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
