RNA-editing-mediated exon evolution
- PMID: 17326827
- PMCID: PMC1852406
- DOI: 10.1186/gb-2007-8-2-r29
RNA-editing-mediated exon evolution
Abstract
Background: Alu retroelements are specific to primates and abundant in the human genome. Through mutations that create functional splice sites within intronic Alus, these elements can become new exons in a process denoted exonization. It was recently shown that Alu elements are also heavily changed by RNA editing in the human genome.
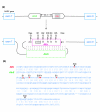
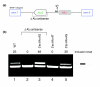
Results: Here we show that the human nuclear prelamin A recognition factor contains a primate-specific Alu-exon that exclusively depends on RNA editing for its exonization. We demonstrate that RNA editing regulates the exonization in a tissue-dependent manner, through both the creation of a functional AG 3' splice site, and alteration of functional exonic splicing enhancers within the exon. Furthermore, a premature stop codon within the Alu-exon is eliminated by an exceptionally efficient RNA editing event. The sequence surrounding this editing site is important not only for editing of that site but also for editing in other neighboring sites as well.
Conclusion: Our results show that the abundant RNA editing of Alu sequences can be recruited as a mechanism supporting the birth of new exons in the human genome.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

