Electron microscopy analysis of viral morphogenesis
- PMID: 17327172
- PMCID: PMC7103132
- DOI: 10.1016/S0091-679X(06)79020-3
Electron microscopy analysis of viral morphogenesis
Abstract
This chapter reviews the use of standard transmission electron microscopy (TEM) of plastic-embedded material, as well as protocols for the immunolabeling of cryosections, in the analysis of viral interactions with cells. It focuses particularly on the assembly of two types of enveloped viruses: (1) the beta herpesvirus—human cytomegalovirus (HCMV), and (2) the primate lentiviruses—the simian and human immunodeficiency viruses (SIV and HIV). The chapter discusses the ways EM is used to identify morphological features of the various stages in the assembly of virus particles, to distinguish immature and mature particles, or to analyze steps involved in the acquisition of lipid membranes by enveloped viruses. In addition, it demonstrates the way immunolabeling allows the quantification of viral components, even in individual virus particles, and comparisons between particles at different locations in the cell or at different stages in viral assembly. Together with the newly developed methods for electron tomography and correlative immunofluorescence studies and EM, huge potential exists to unravel more details about virus assembly in the near future.
Figures
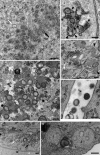
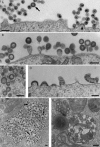
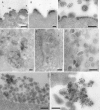
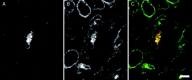
Similar articles
-
Virus morphogenesis in the cell: methods and observations.Subcell Biochem. 2013;68:417-40. doi: 10.1007/978-94-007-6552-8_14. Subcell Biochem. 2013. PMID: 23737060 Free PMC article. Review.
-
Theoretical aspects of structure and assembly of viral envelops.Curr Top Microbiol Immunol. 1975;70:1-30. doi: 10.1007/978-3-642-66101-3_1. Curr Top Microbiol Immunol. 1975. PMID: 808396 Review. No abstract available.
-
Electron microscopy: essentials for viral structure, morphogenesis and rapid diagnosis.Sci China Life Sci. 2013 May;56(5):421-30. doi: 10.1007/s11427-013-4476-2. Epub 2013 May 1. Sci China Life Sci. 2013. PMID: 23633074 Free PMC article. Review.
-
Viral life cycles captured in three-dimensions with electron microscopy tomography.Curr Opin Virol. 2011 Aug;1(2):125-33. doi: 10.1016/j.coviro.2011.06.008. Curr Opin Virol. 2011. PMID: 21887207 Free PMC article. Review.
-
Electron microscopy of rapid identification of animal viruses in hematoxylin-eosin sections.Can J Comp Med. 1977 Oct;41(4):416-9. Can J Comp Med. 1977. PMID: 72592 Free PMC article.
Cited by
-
Expression, purification, and functional characterization of soluble recombinant full-length simian immunodeficiency virus (SIV) Pr55Gag.Heliyon. 2023 Jan 10;9(1):e12892. doi: 10.1016/j.heliyon.2023.e12892. eCollection 2023 Jan. Heliyon. 2023. PMID: 36685375 Free PMC article.
-
Viral detection by electron microscopy: past, present and future.Biol Cell. 2008 Aug;100(8):491-501. doi: 10.1042/BC20070173. Biol Cell. 2008. PMID: 18627353 Free PMC article.
-
Adenoviral infection in 5 red-tailed hawks and a broad-winged hawk.J Vet Diagn Invest. 2022 Sep;34(5):796-805. doi: 10.1177/10406387221105240. Epub 2022 Jun 27. J Vet Diagn Invest. 2022. PMID: 35762098 Free PMC article.
-
The intracellular plasma membrane-connected compartment in the assembly of HIV-1 in human macrophages.BMC Biol. 2016 Jun 23;14:50. doi: 10.1186/s12915-016-0272-3. BMC Biol. 2016. PMID: 27338237 Free PMC article.
-
The viral replication organelles within cells studied by electron microscopy.Adv Virus Res. 2019;105:1-33. doi: 10.1016/bs.aivir.2019.07.005. Epub 2019 Aug 19. Adv Virus Res. 2019. PMID: 31522702 Free PMC article.
References
-
- Briggs J.A., Grunewald K., Glass B., Forster F., Krausslich H.G., Fuller S.D. The mechanism of HIV‐1 core assembly: Insights from three‐dimensional reconstructions of authentic virions. Structure. 2006;14:15–20. - PubMed
-
- Briggs J.A., Simon M.N., Gross I., Krausslich H.G., Fuller S.D., Vogt V.M., Johnson M.C. The stoichiometry of Gag protein in HIV‐1. Nat. Struct. Mol. Biol. 2004;11:672–675. - PubMed
-
- Britt W.J., Boppana S. Human cytomegalovirus virion proteins. Hum. Immunol. 2004;65:395–402. - PubMed
-
- Britt W.J., Mach M. Human cytomegalovirus glycoproteins. Intervirology. 1996;39:401–412. - PubMed
-
- Canto‐Nogues C., Hockley D., Grief C., Ranjbar S., Bootman J., Almond N., Herrera I. Ultrastructural localization of the RNA of immunodeficiency viruses using electron microscopy in situ hybridization and in vitro infected lymphocytes. Micron. 2001;32:579–589. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

