Genome-wide analysis of neuroblastomas using high-density single nucleotide polymorphism arrays
- PMID: 17327916
- PMCID: PMC1797488
- DOI: 10.1371/journal.pone.0000255
Genome-wide analysis of neuroblastomas using high-density single nucleotide polymorphism arrays
Abstract
Background: Neuroblastomas are characterized by chromosomal alterations with biological and clinical significance. We analyzed paired blood and primary tumor samples from 22 children with high-risk neuroblastoma for loss of heterozygosity (LOH) and DNA copy number change using the Affymetrix 10K single nucleotide polymorphism (SNP) array.
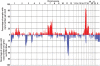
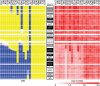
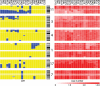
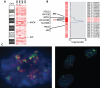
Findings: Multiple areas of LOH and copy number gain were seen. The most commonly observed area of LOH was on chromosome arm 11q (15/22 samples; 68%). Chromosome 11q LOH was highly associated with occurrence of chromosome 3p LOH: 9 of the 15 samples with 11q LOH had concomitant 3p LOH (P = 0.016). Chromosome 1p LOH was seen in one-third of cases. LOH events on chromosomes 11q and 1p were generally accompanied by copy number loss, indicating hemizygous deletion within these regions. The one exception was on chromosome 11p, where LOH in all four cases was accompanied by normal copy number or diploidy, implying uniparental disomy. Gain of copy number was most frequently observed on chromosome arm 17q (21/22 samples; 95%) and was associated with allelic imbalance in six samples. Amplification of MYCN was also noted, and also amplification of a second gene, ALK, in a single case.
Conclusions: This analysis demonstrates the power of SNP arrays for high-resolution determination of LOH and DNA copy number change in neuroblastoma, a tumor in which specific allelic changes drive clinical outcome and selection of therapy.
Conflict of interest statement
Figures

References
-
- Wang DG, Fan JB, Siao CJ, Berno A, Young P, et al. Large-scale identification, mapping, and genotyping of single-nucleotide polymorphisms in the human genome. Science. 1998;280:1077–1082. - PubMed
-
- Zhao X, Li C, Paez JG, Chin K, Janne PA, et al. An integrated view of copy number and allelic alterations in the cancer genome using single nucleotide polymorphism arrays. Cancer Res. 2004;64:3060–3071. - PubMed
-
- Janne PA, Li C, Zhao X, Girard L, Chen TH, et al. High-resolution single-nucleotide polymorphism array and clustering analysis of loss of heterozygosity in human lung cancer cell lines. Oncogene. 2004;23:2716–2726. - PubMed
-
- Lindblad-Toh K, Tanenbaum DM, Daly MJ, Winchester E, Lui WO, et al. Loss-of-heterozygosity analysis of small-cell lung carcinomas using single-nucleotide polymorphism arrays. Nat Biotechnol. 2000;18:1001–1005. - PubMed
-
- Lieberfarb ME, Lin M, Lechpammer M, Li C, Tanenbaum DM, et al. Genome-wide loss of heterozygosity analysis from laser capture microdissected prostate cancer using single nucleotide polymorphic allele (SNP) arrays and a novel bioinformatics platform dChipSNP. Cancer Res. 2003;63:4781–4785. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

