The coding/non-coding overlapping architecture of the gene encoding the Drosophila pseudouridine synthase
- PMID: 17328797
- PMCID: PMC1821038
- DOI: 10.1186/1471-2199-8-15
The coding/non-coding overlapping architecture of the gene encoding the Drosophila pseudouridine synthase
Abstract
Background: In eukaryotic cells, each molecule of H/ACA small nucleolar RNA (snoRNA) assembles with four evolutionarily conserved core proteins to compose a specific ribonucleoprotein particle. One of the four core components has pseudouridine synthase activity and catalyzes the conversion of a selected uridine to pseudouridine. Members of the pseudouridine synthase family are highly conserved. In addition to catalyzing pseudouridylation of target RNAs, they carry out a variety of essential functions related to ribosome biogenesis and, in mammals, to telomere maintenance. To investigate further the molecular mechanisms underlying the expression of pseudouridine synthase genes, we analyzed the transcriptional activity of the Drosophila member of this family in great detail.
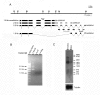
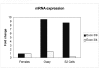
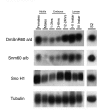
Results: The Drosophila gene for pseudouridine synthase, minifly/Nop60b (mfl), encodes two novel mRNAs ending at a downstream poly(A) site. One species is characterized only by an extended 3'-untranslated region (3'UTR), while a minor mRNA encodes a variant protein that represents the first example of an alternative subform described for any member of the family to date. The rare spliced variant is detected mainly in females and is predicted to have distinct functional properties. We also report that a cluster comprising four isoforms of a C/D box snoRNA and two highly related copies of a small ncRNA gene of unknown function is intron-encoded at the gene-variable 3'UTRs. Because this arrangement, the alternative 3' ends allow mfl not only to produce two distinct protein subforms, but also to release different ncRNAs. Intriguingly, accumulation of all these intron-encoded RNAs was found to be sex-biased and quantitatively modulated throughout development and, within the ovaries, the ncRNAs of unknown function were found not ubiquitously expressed.
Conclusion: Our results expand the repertoire of coding/non-coding transcripts derived from the gene encoding Drosophila pseudouridine synthase. This gene exhibits a complex and interlaced organization, and its genetic information may be expressed as different protein subforms and/or ncRNAs that may potentially contribute to its biological functions.
Figures

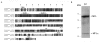

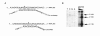


References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

