Development of Sindbis viruses encoding nsP2/GFP chimeric proteins and their application for studying nsP2 functioning
- PMID: 17329335
- PMCID: PMC1900196
- DOI: 10.1128/JVI.02746-06
Development of Sindbis viruses encoding nsP2/GFP chimeric proteins and their application for studying nsP2 functioning
Abstract
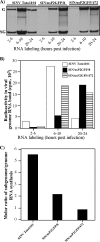
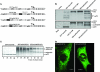
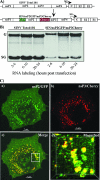
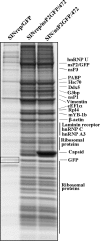
Sindbis virus (SINV) is one of almost 30 currently known alphaviruses. In infected cells, it produces only a few proteins that function in virus replication and interfere with the development of the antiviral response. One of the viral nonstructural proteins, nsP2, not only exhibits protease and RNA helicase activities that are directly involved in viral RNA replication but also plays critical roles in the development of transcriptional and translational shutoffs in the SINV-infected cells. These multiple activities of nsP2 complicate investigations of this protein's functions and further understanding of its structure. Using a transposon-based approach, we generated a cDNA library of SINV genomes with a green fluorescent protein (GFP) gene randomly inserted into nsP2 and identified a number of sites that can be used for GFP cloning without a strong effect on virus replication. Recombinant SIN viruses encoding nsP2/GFP chimeric protein were capable of growth in tissue culture and interfering with cellular functions. SINV, expressing GFP in the nsP2, was used to isolate nsP2-specific protein complexes formed in the cytoplasm of the infected cells. These complexes contained viral nsPs, all of the cellular proteins that we previously coisolated with SINV nsP3, and some additional protein factors that were not found before in detectable concentrations. The random insertion library-based approach, followed by the selection of the viable variants expressing heterologous proteins, can be applied for mapping the domain structure of the viral nonstructural and structural proteins, cloning of peptide tags for isolation of the protein-specific complexes, and studying their formation by using live-cell imaging. This approach may also be applicable to presentation of additional antigens and retargeting of viruses to new receptors.
Figures




References
-
- Agapov, E. A., K. E. Reed, and C. M. Rice. 1998. Use of the vaccinia virus expression system for the study of HCV protein processing, p. 303-314. In J. Y. N. Lau (ed.), Methods in molecular medicine, vol. 19. Humana Press, New York, NY. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

