The Arabidopsis thaliana homolog of yeast BRE1 has a function in cell cycle regulation during early leaf and root growth
- PMID: 17329565
- PMCID: PMC1867331
- DOI: 10.1105/tpc.106.041319
The Arabidopsis thaliana homolog of yeast BRE1 has a function in cell cycle regulation during early leaf and root growth
Abstract
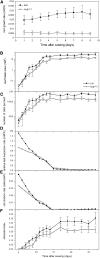
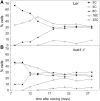
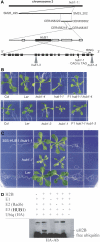
Chromatin modification and transcriptional activation are novel roles for E3 ubiquitin ligase proteins that have been mainly associated with ubiquitin-dependent proteolysis. We identified HISTONE MONOUBIQUITINATION1 (HUB1) (and its homolog HUB2) in Arabidopsis thaliana as RING E3 ligase proteins with a function in organ growth. We show that HUB1 is a functional homolog of the human and yeast BRE1 proteins because it monoubiquitinated histone H2B in an in vitro assay. Hub knockdown mutants had pale leaf coloration, modified leaf shape, reduced rosette biomass, and inhibited primary root growth. One of the alleles had been designated previously as ang4-1. Kinematic analysis of leaf and root growth together with flow cytometry revealed defects in cell cycle activities. The hub1-1 (ang4-1) mutation increased cell cycle duration in young leaves and caused an early entry into the endocycles. Transcript profiling of shoot apical tissues of hub1-1 (ang4-1) indicated that key regulators of the G2-to-M transition were misexpressed. Based on the mutant characterization, we postulate that HUB1 mediates gene activation and cell cycle regulation probably through chromatin modifications.
Figures



Comment in
-
Two tales of chromatin remodeling converge on HUB1.Plant Cell. 2007 Feb;19(2):391-3. doi: 10.1105/tpc.107.190210. Plant Cell. 2007. PMID: 17389225 Free PMC article. No abstract available.
Similar articles
-
The absence of histone H2B monoubiquitination in the Arabidopsis hub1 (rdo4) mutant reveals a role for chromatin remodeling in seed dormancy.Plant Cell. 2007 Feb;19(2):433-44. doi: 10.1105/tpc.106.049221. Epub 2007 Feb 28. Plant Cell. 2007. PMID: 17329563 Free PMC article.
-
Two tales of chromatin remodeling converge on HUB1.Plant Cell. 2007 Feb;19(2):391-3. doi: 10.1105/tpc.107.190210. Plant Cell. 2007. PMID: 17389225 Free PMC article. No abstract available.
-
Histone H2B monoubiquitination is involved in the regulation of cutin and wax composition in Arabidopsis thaliana.Plant Cell Physiol. 2014 Feb;55(2):455-66. doi: 10.1093/pcp/pct182. Epub 2013 Dec 5. Plant Cell Physiol. 2014. PMID: 24319075
-
Molecular networks regulating cell division during Arabidopsis leaf growth.J Exp Bot. 2020 Apr 23;71(8):2365-2378. doi: 10.1093/jxb/erz522. J Exp Bot. 2020. PMID: 31748815 Free PMC article. Review.
-
Coordination of cell proliferation and cell expansion in the control of leaf size in Arabidopsis thaliana.J Plant Res. 2006 Jan;119(1):37-42. doi: 10.1007/s10265-005-0232-4. Epub 2005 Nov 12. J Plant Res. 2006. PMID: 16284709 Review.
Cited by
-
OsHUB1 and OsHUB2 interact with SPIN6 and form homo- and hetero-dimers in rice.Plant Signal Behav. 2015;10(8):e1039212. doi: 10.1080/15592324.2015.1039212. Plant Signal Behav. 2015. PMID: 25955387 Free PMC article.
-
RING Zinc Finger Proteins in Plant Abiotic Stress Tolerance.Front Plant Sci. 2022 Apr 14;13:877011. doi: 10.3389/fpls.2022.877011. eCollection 2022. Front Plant Sci. 2022. PMID: 35498666 Free PMC article. Review.
-
The Ubiquitin Conjugating Enzyme: An Important Ubiquitin Transfer Platform in Ubiquitin-Proteasome System.Int J Mol Sci. 2020 Apr 21;21(8):2894. doi: 10.3390/ijms21082894. Int J Mol Sci. 2020. PMID: 32326224 Free PMC article. Review.
-
Nonredundant requirement for multiple histone modifications for the early anaphase release of the mitotic exit regulator Cdc14 from nucleolar chromatin.PLoS Genet. 2009 Aug;5(8):e1000588. doi: 10.1371/journal.pgen.1000588. Epub 2009 Aug 7. PLoS Genet. 2009. PMID: 19662160 Free PMC article.
-
The Arabidopsis cell division cycle.Arabidopsis Book. 2009;7:e0120. doi: 10.1199/tab.0120. Epub 2009 Mar 20. Arabidopsis Book. 2009. PMID: 22303246 Free PMC article.
References
-
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. - PubMed
-
- Beemster, G.T.S., De Veylder, L., Vercruysse, S., West, G., Rombaut, D., Van Hummelen, P., Galichet, A., Gruissem, W., Inzé, D., and Vuylsteke, M. (2005). Genome-wide analysis of gene expression profiles associated with cell cycle transitions in growing organs of Arabidopsis. Plant Physiol. 138 734–743. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

