Conservation of Toll-Like Receptor Signaling Pathways in Teleost Fish
- PMID: 17330145
- PMCID: PMC1524722
- DOI: 10.1016/j.cbd.2005.07.003
Conservation of Toll-Like Receptor Signaling Pathways in Teleost Fish
Abstract
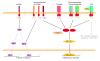
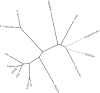
In mammals, Toll-like receptors (TLR) recognize ligands, including pathogen-associated molecular patterns (PAMPs), and respond with ligand-specific induction of genes. In this study, we establish evolutionary conservation in teleost fish of key components of the TLR-signaling pathway that act as switches for differential gene induction, including MYD88, TIRAP, TRIF, TRAF6, IRF3, and IRF7. We further explore this conservation with a molecular phylogenetic analysis of MYD88. To the extent that current genomic analysis can establish, each vertebrate has one ortholog to each of these genes. For molecular tree construction and phylogeny inference, we demonstrate a methodology for including genes with only partial primary sequences without disrupting the topology provided by the high-confidence full-length sequences. Conservation of the TLR-signaling molecules suggests that the basic program of gene regulation by the TLR-signaling pathway is conserved across vertebrates. To test this hypothesis, leukocytes from a model fish, rainbow trout (Oncorhynchus mykiss), were stimulated with known mammalian TLR agonists including: diacylated and triacylated forms of lipoprotein, flagellin, two forms of LPS, synthetic double-stranded RNA, and two imidazoquinoline compounds (loxoribine and R848). Trout leukocytes responded in vitro to a number of these agonists with distinct patterns of cytokine expression that correspond to mammalian responses. Our results support the key prediction from our phylogenetic analyses that strong selective pressure of pathogenic microbes has preserved both TLR recognition and signaling functions during vertebrate evolution.
Figures

References
-
- Aderem A, Smith KD. A systems approach to dissecting immunity and inflammation. Sem Immunol. 2004;16:55–67. - PubMed
-
- Akira S. Toll-like receptor signaling. J Biol Chem. 2003;278:38105–38108. - PubMed
-
- Akira S, Yamamoto M, Takeda K. Role of adapters in Toll-like receptor signalling. Biochem Soc Trans. 2003;31:637–642. - PubMed
-
- Allendorf, F.W., Thorgaard, G. 1984. Tetraploidy and the evolution of salmonid fishes. In: Turner, B.J. (Ed.) The Evolutionary Genetics of Fishes, Plenum Press, New York, pp. 1–53
-
- Aparicio S, Chapman J, Stupka E, Putnam N, Chia J, Dehal P, Christoffels A, Rash S, Hoon S, Smit A, Gelpke MDS, Roach J, Oh T, Ho IY, Wong M, Detter C, Verhoef F, Predki P, Tay A, Lucas S, Richardson P, Smith SF, Clark MS, Edwards YJK, Doggett N, Zharkikh A, Tavtigian SV, Pruss D, Barnstead M, Evans C, Baden H, Powell J, Glusman G, Rowen L, Hood L, Tan YH, Elgar G, Hawkins T, Venkatesh B, Rokhsar D, Brenner S. Whole-genome shotgun assembly and analysis of the genome of Fugu rubripes. Science. 2002;297:1301–1310. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

