Cauliflower mosaic virus: a 420 subunit (T = 7), multilayer structure
- PMID: 1733107
- PMCID: PMC4167691
- DOI: 10.1016/0042-6822(92)90032-k
Cauliflower mosaic virus: a 420 subunit (T = 7), multilayer structure
Abstract
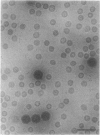
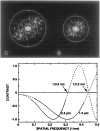
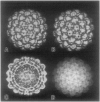
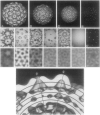
The structures of the Cabb-B and CM1841 strains of cauliflower mosaic virus (CaMV) have been solved to about 3 nm resolution from unstained, frozen-hydrated samples that were examined with low-irradiation cryo-electron microscopy and three-dimensional image reconstruction procedures. CaMV is highly susceptible to distortions. Spherical particles, with a maximum diameter of 53.8 nm, are composed of three concentric layers (I-III) of solvent-excluded density that surround a large, solvent-filled cavity (approximately 27 nm dia). The outermost layer (I) contains 72 capsomeric morphological units, with 12 pentavalent pentamers and 60 hexavalent hexamers for a total of 420 subunits (37-42 kDa each) arranged with T = 7 icosahedral symmetry. CaMV is the first example of a T = 7 virus that obeys the rules of stoichiometry proposed for isometric viruses by Caspar and Klug (1962, Cold Spring Harb. Symp. Quant. Biol. 27, 1-24), although the hexameric capsomers exhibit marked departure from the regular sixfold symmetry expected for a structure in which the capsid protein subunits are quasi-equivalently related. The double-stranded DNA genome is distributed in layers II and III along with a portion of the viral protein. The CaMV reconstructions are consistent with the model based on neutron diffraction studies (Kruse et al., 1987, Virology 159, 166-168) and, together, these structural models are discussed in relation to a replication-assembly model (Hull et al., 1987, J. Cell Sci. (Suppl.) 7, 213-229). Remarkable agreement between the reconstructions of CaMV Cabb-B and CM1841 suggests that other strains of CaMV adopt the same basic structure.
Figures




References
-
- Al Ani R, Pfeiffer P, Lebeurier G, Hirth L. The structure of cauliflower mosaic virus. I pH-induced structural changes Virology. 1979a;93:175–187. - PubMed
-
- Al Ani R, Pfeiffer P, Lebeurier G. The structure of cauliflower mosaic virus. II. Identity and location of the viral polypeptides. Virology. 1979b;93:188–197. - PubMed
