Notch signaling is essential for ventricular chamber development
- PMID: 17336907
- PMCID: PMC2746361
- DOI: 10.1016/j.devcel.2006.12.011
Notch signaling is essential for ventricular chamber development
Abstract
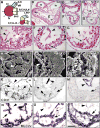
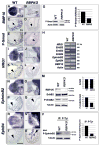
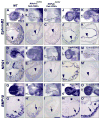
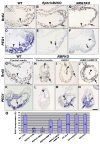
Ventricular chamber morphogenesis, first manifested by trabeculae formation, is crucial for cardiac function and embryonic viability and depends on cellular interactions between the endocardium and myocardium. We show that ventricular Notch1 activity is highest at presumptive trabecular endocardium. RBPJk and Notch1 mutants show impaired trabeculation and marker expression, attenuated EphrinB2, NRG1, and BMP10 expression and signaling, and decreased myocardial proliferation. Functional and molecular analyses show that Notch inhibition prevents EphrinB2 expression, and that EphrinB2 is a direct Notch target acting upstream of NRG1 in the ventricles. However, BMP10 levels are found to be independent of both EphrinB2 and NRG1 during trabeculation. Accordingly, exogenous BMP10 rescues the myocardial proliferative defect of in vitro-cultured RBPJk mutants, while exogenous NRG1 rescues differentiation in parallel. We suggest that during trabeculation Notch independently regulates cardiomyocyte proliferation and differentiation, two exquisitely balanced processes whose perturbation may result in congenital heart disease.
Figures


References
-
- Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–776. - PubMed
-
- Bailey AM, Posakony JW. Suppressor of hairless directly activates transcription of enhancer of split complex genes in response to Notch receptor activity. Genes Dev. 1995;9:2609–2622. - PubMed
-
- Ben-Shachar G, Arcilla RA, Lucas RV, Manasek FJ. Ventricular trabeculations in the chick embryo heart and their contribution to ventricular and muscular septal development. Circ Res. 1985;57:759–766. - PubMed
-
- Conlon RA, Reaume AG, Rossant J. Notch1 is required for the coordinate segmentation of somites. Development. 1995;121:1533–1545. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

