Human mitochondrial ribosomal protein MRPL12 interacts directly with mitochondrial RNA polymerase to modulate mitochondrial gene expression
- PMID: 17337445
- PMCID: PMC2606046
- DOI: 10.1074/jbc.M700461200
Human mitochondrial ribosomal protein MRPL12 interacts directly with mitochondrial RNA polymerase to modulate mitochondrial gene expression
Abstract
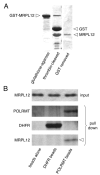
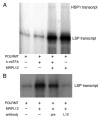
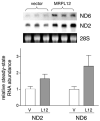
The core human mitochondrial transcription machinery comprises a single subunit bacteriophage-related RNA polymerase, POLRMT, the high mobility group box DNA-binding protein h-mtTFA/TFAM, and two transcriptional co-activator proteins, h-mtTFB1 and h-mtTFB2 that also have rRNA methyltransferase activity. Recapitulation of specific initiation of transcription in vitro can be achieved by a complex of POL-RMT, h-mtTFA, and either h-mtTFB1 or h-mtTFB2. However, the nature of mitochondrial transcription complexes in vivo and the potential involvement of additional proteins in the transcription process in human mitochondria have not been extensively investigated. In Saccharomyces cerevisiae, transcription and translation are physically coupled via the formation of a multiprotein complex nucleated by the binding of Nam1p to the amino-terminal domain of mtRNA polymerase (Rpo41p). This model system paradigm led us to search for proteins that interact with POLRMT to regulate mitochondrial gene expression in humans. Using an affinity capture strategy to identify POL-RMT-binding proteins, we identified mitochondrial ribosomal protein L7/L12 (MRPL12) as a protein in HeLa mitochondrial extracts that interacts specifically with POLRMT in vitro. Purified recombinant MRPL12 binds to POLRMT and stimulates mitochondrial transcription activity in vitro, demonstrating that this interaction is both direct and functional. Finally, from HeLa cells that overexpress FLAG epitope-tagged MRPL12, increased steady-state levels of mtDNA-encoded transcripts are observed and MRPL12-POLRMT complexes can be co-immunoprecipitated, providing strong evidence that this interaction enhances mitochondrial transcription or RNA stability in vivo. We speculate that the MRPL12 interaction with POLRMT is likely part of a novel regulatory mechanism that coordinates mitochondrial transcription with translation and/or ribosome biogenesis during human mitochondrial gene expression.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

