PlanTAPDB, a phylogeny-based resource of plant transcription-associated proteins
- PMID: 17337525
- PMCID: PMC1851845
- DOI: 10.1104/pp.107.095760
PlanTAPDB, a phylogeny-based resource of plant transcription-associated proteins
Abstract
Diversification of transcription-associated protein (TAP) families during land plant evolution is a key process yielding increased complexity of plant life. Understanding the evolutionary relationships between these genes is crucial to gain insight into plant evolution. We have determined a substantial set of TAPs that are focused on, but not limited to, land plants using PSI-BLAST searches and subsequent filtering and clustering steps. Phylogenies were created in an automated way using a combination of distance and maximum likelihood methods. Comparison of the data to previously published work confirmed their accuracy and usefulness for the majority of gene families. Evidence is presented that the flowering plant apical stem cell regulator WUSCHEL evolved from an ancestral homeobox gene that was already present after the water-to-land transition. The presence of distinct expanded gene families, such as COP1 and HIT in moss, is discussed within the evolutionary backdrop. Comparative analyses revealed that almost all angiosperm transcription factor families were already present in the earliest land plants, whereas many are missing among unicellular algae. A global analysis not only of transcription factors but also of transcriptional regulators and novel putative families is presented. A wealth of data about plant TAP families and all data accrued throughout their automated detection and analysis are made available via the PlanTAPDB Web interface. Evolutionary relationships of these genes are readily accessible to the nonexpert at a mouse-click. Initial analyses of selected gene families revealed that PlanTAPDB can easily be exerted for knowledge discovery.
Figures

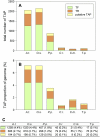




References
-
- Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B 57 289–300
-
- Bierfreund NM, Tintelnot S, Reski R, Decker EL (2004) Loss of GH3 function does not affect phytochrome-mediated development in a moss, Physcomitrella patens. J Plant Physiol 161 823–835 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

