Dynamics of genomic-library enrichment and identification of solvent tolerance genes for Clostridium acetobutylicum
- PMID: 17337545
- PMCID: PMC1892849
- DOI: 10.1128/AEM.02296-06
Dynamics of genomic-library enrichment and identification of solvent tolerance genes for Clostridium acetobutylicum
Abstract
A Clostridium acetobutylicum ATCC 824 genomic library was constructed using randomly sheared DNA. Library inserts conferring increased tolerance to 1-butanol were isolated using two protocols. Protocol I utilized a single round of butanol challenges in batch culture, while protocol II, which gave clearly superior outcomes, was based on the serial transfer of stationary-phase cultures into progressively higher butanol concentrations. DNA microarray analysis made a high-resolution assessment of the dynamic process of library enrichment possible for the first time. Protocol I yielded a library insert containing the entire coding region of the gene CAC0003 (which codes for a protein of unknown function) but also several DNA fragments containing promoter regions. Protocol II enabled the successful identification of DNA fragments containing several intact genes conferring preferential growth under conditions of butanol stress. Since expression using the employed library is possible only from natural promoters, among the enriched genes, we identified 16 genes that constitute the first cistron of a transcriptional unit. These genes include four transcriptional regulators (CAC0977, CAC1463, CAC1869, and CAC2495). After subcloning plasmids carrying the CAC0003 and CAC1869 genes, strains 824(pCAC0003) and 824(pCAC1869) exhibited 13% and an 81% increases, respectively, in butanol tolerance relative to the plasmid control strain. 824(pCAC1869) consistently grew to higher cell densities in challenged and unchallenged cultures and exhibited prolonged metabolism. Our serial enrichment approach provided a more detailed understanding of the dynamic process of library enrichment under conditions of selective growth. Further characterization of the genes identified in this study will likely enhance our understanding of the complex phenotype of solvent tolerance.
Figures
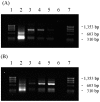
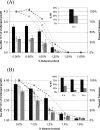
 ), 824(pIMP1) (
), 824(pIMP1) ( ), and 824(pCAC0003) (□). Growth measurements were used to calculate the percent tolerance at each challenge level for 824(pCAC1869) (□), 824(pIMP1) (○), and 824(pCAC0003) (▵). (A) Inoculation of a dilution series of butanol in CGM. (B) Butanol challenge of actively growing cultures (A600 of ∼1.0). Growth data for 12 h postchallenge are displayed. Inset graphs show the percent relative tolerance (% RT) to butanol for library strains relative to the pIMP1 control strain. See the text for details.
), and 824(pCAC0003) (□). Growth measurements were used to calculate the percent tolerance at each challenge level for 824(pCAC1869) (□), 824(pIMP1) (○), and 824(pCAC0003) (▵). (A) Inoculation of a dilution series of butanol in CGM. (B) Butanol challenge of actively growing cultures (A600 of ∼1.0). Growth data for 12 h postchallenge are displayed. Inset graphs show the percent relative tolerance (% RT) to butanol for library strains relative to the pIMP1 control strain. See the text for details.Similar articles
-
A transcriptional study of acidogenic chemostat cells of Clostridium acetobutylicum--solvent stress caused by a transient n-butanol pulse.J Biotechnol. 2012 Oct 31;161(3):354-65. doi: 10.1016/j.jbiotec.2012.03.027. Epub 2012 Apr 15. J Biotechnol. 2012. PMID: 22537853
-
A genomic-library based discovery of a novel, possibly synthetic, acid-tolerance mechanism in Clostridium acetobutylicum involving non-coding RNAs and ribosomal RNA processing.Metab Eng. 2010 May;12(3):268-81. doi: 10.1016/j.ymben.2009.12.004. Epub 2010 Jan 6. Metab Eng. 2010. PMID: 20060060 Free PMC article.
-
Engineering Clostridium acetobutylicum with a histidine kinase knockout for enhanced n-butanol tolerance and production.Appl Microbiol Biotechnol. 2015 Jan;99(2):1011-22. doi: 10.1007/s00253-014-6249-7. Epub 2014 Dec 5. Appl Microbiol Biotechnol. 2015. PMID: 25472438
-
Comparative genomic and proteomic analyses of Clostridium acetobutylicum Rh8 and its parent strain DSM 1731 revealed new understandings on butanol tolerance.Biochem Biophys Res Commun. 2014 Aug 8;450(4):1612-8. doi: 10.1016/j.bbrc.2014.07.052. Epub 2014 Jul 17. Biochem Biophys Res Commun. 2014. PMID: 25044112
-
Identification and characterization of two functionally unknown genes involved in butanol tolerance of Clostridium acetobutylicum.PLoS One. 2012;7(6):e38815. doi: 10.1371/journal.pone.0038815. Epub 2012 Jun 29. PLoS One. 2012. PMID: 22768047 Free PMC article.
Cited by
-
Genetic determinants for n-butanol tolerance in evolved Escherichia coli mutants: cross adaptation and antagonistic pleiotropy between n-butanol and other stressors.Appl Environ Microbiol. 2013 Sep;79(17):5313-20. doi: 10.1128/AEM.01703-13. Epub 2013 Jun 28. Appl Environ Microbiol. 2013. PMID: 23811509 Free PMC article.
-
Evolution combined with genomic study elucidates genetic bases of isobutanol tolerance in Escherichia coli.Microb Cell Fact. 2011 Mar 25;10:18. doi: 10.1186/1475-2859-10-18. Microb Cell Fact. 2011. PMID: 21435272 Free PMC article.
-
Coexisting/Coexpressing Genomic Libraries (CoGeL) identify interactions among distantly located genetic loci for developing complex microbial phenotypes.Nucleic Acids Res. 2011 Dec;39(22):e152. doi: 10.1093/nar/gkr817. Epub 2011 Oct 5. Nucleic Acids Res. 2011. PMID: 21976725 Free PMC article.
-
Investigation of availability of a high throughput screening method for predicting butanol solvent -producing ability of Clostridium beijerinckii.BMC Microbiol. 2016 Jul 22;16(1):160. doi: 10.1186/s12866-016-0776-6. BMC Microbiol. 2016. PMID: 27448996 Free PMC article.
-
Strain and bioprocess improvement of a thermophilic anaerobe for the production of ethanol from wood.Biotechnol Biofuels. 2016 Jun 16;9:125. doi: 10.1186/s13068-016-0536-8. eCollection 2016. Biotechnol Biofuels. 2016. PMID: 27313661 Free PMC article.
References
-
- Alper, H., J. Moxley, E. Nevoigt, G. R. Fink, and G. Stephanopoulos. 2006. Engineering yeast transcription machinery for improved ethanol tolerance and production. Science 314:1565-1568. - PubMed
-
- Alsaker, K. V. 2006. Microarray-based transcriptional analyses of stationary phase phenomena and stress responses in Clostridium acetobutylicum. Ph.D. thesis. Northwestern University, Evanston, IL.
-
- Alsaker, K. V., C. J. Paredes, and E. T. Papoutsakis. 2005. Design, optimization and validation of genomic DNA microarrays for examining the Clostridium acetobutylicum transcriptome. Biotechnol. Bioprocess Eng. 10:432-443.
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

