The alternative sigma factor sigma B and the virulence gene regulator PrfA both regulate transcription of Listeria monocytogenes internalins
- PMID: 17337550
- PMCID: PMC1892873
- DOI: 10.1128/AEM.02664-06
The alternative sigma factor sigma B and the virulence gene regulator PrfA both regulate transcription of Listeria monocytogenes internalins
Abstract
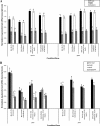
Some Listeria monocytogenes internalins are recognized as contributing to invasion of mammalian tissue culture cells. While PrfA is well established as a positive regulator of L. monocytogenes virulence gene expression, the stress-responsive sigma(B) has been recognized only recently as contributing to expression of virulence genes, including some that encode internalins. To measure the relative contributions of PrfA and sigma(B) to internalin gene transcription, we used reverse transcription-PCR to quantify transcript levels for eight internalin genes (inlA, inlB, inlC, inlC2, inlD, inlE, inlF, and inlG) in L. monocytogenes 10403S and in isogenic Delta prfA, Delta sigB, and Delta sigB Delta prfA strains. Strains were grown under defined conditions to produce (i) active PrfA, (ii) active sigma(B) and active PrfA, (iii) inactive PrfA, and (iv) active sigma(B) and inactive PrfA. Under the conditions tested, sigma(B) and PrfA contributed differentially to the expression of the various internalins such that (i) both sigma(B) and PrfA contributed to inlA and inlB transcription, (ii) only PrfA contributed to inlC transcription, (iii) only sigma(B) contributed to inlC2 and inlD transcription, and (iv) neither sigma(B) nor PrfA contributed to inlF and inlG transcription. inlE transcript levels were undetectable. The important role for sigma(B) in regulating expression of L. monocytogenes internalins suggests that exposure of this organism to environmental stress conditions, such as those encountered in the gastrointestinal tract, may activate internalin transcription. Interplay between sigma(B) and PrfA also appears to be critical for regulating transcription of some virulence genes, including inlA, inlB, and prfA.
Figures


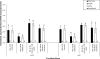

Similar articles
-
Differential regulation of Listeria monocytogenes internalin and internalin-like genes by sigmaB and PrfA as revealed by subgenomic microarray analyses.Foodborne Pathog Dis. 2008 Aug;5(4):417-35. doi: 10.1089/fpd.2008.0085. Foodborne Pathog Dis. 2008. PMID: 18713061 Free PMC article.
-
Contributions of six lineage-specific internalin-like genes to invasion efficiency of Listeria monocytogenes.Foodborne Pathog Dis. 2009 Jan-Feb;6(1):57-70. doi: 10.1089/fpd.2008.0140. Foodborne Pathog Dis. 2009. PMID: 19014275
-
Temperature-dependent expression of Listeria monocytogenes internalin and internalin-like genes suggests functional diversity of these proteins among the listeriae.Appl Environ Microbiol. 2007 May;73(9):2806-14. doi: 10.1128/AEM.02923-06. Epub 2007 Mar 2. Appl Environ Microbiol. 2007. PMID: 17337561 Free PMC article.
-
Cross Talk between SigB and PrfA in Listeria monocytogenes Facilitates Transitions between Extra- and Intracellular Environments.Microbiol Mol Biol Rev. 2019 Sep 4;83(4):e00034-19. doi: 10.1128/MMBR.00034-19. Print 2019 Nov 20. Microbiol Mol Biol Rev. 2019. PMID: 31484692 Free PMC article. Review.
-
Flick of a switch: regulatory mechanisms allowing Listeria monocytogenes to transition from a saprophyte to a killer.Microbiology (Reading). 2019 Aug;165(8):819-833. doi: 10.1099/mic.0.000808. Epub 2019 May 20. Microbiology (Reading). 2019. PMID: 31107205 Review.
Cited by
-
Genes Associated with Desiccation and Osmotic Stress in Listeria monocytogenes as Revealed by Insertional Mutagenesis.Appl Environ Microbiol. 2015 Aug 15;81(16):5350-62. doi: 10.1128/AEM.01134-15. Epub 2015 May 29. Appl Environ Microbiol. 2015. PMID: 26025900 Free PMC article.
-
Listeria monocytogenes sigmaB modulates PrfA-mediated virulence factor expression.Infect Immun. 2009 May;77(5):2113-24. doi: 10.1128/IAI.01205-08. Epub 2009 Mar 2. Infect Immun. 2009. PMID: 19255187 Free PMC article.
-
The PlcR virulence regulon of Bacillus cereus.PLoS One. 2008 Jul 30;3(7):e2793. doi: 10.1371/journal.pone.0002793. PLoS One. 2008. PMID: 18665214 Free PMC article.
-
Biofilm-isolated Listeria monocytogenes exhibits reduced systemic dissemination at the early (12-24 h) stage of infection in a mouse model.NPJ Biofilms Microbiomes. 2021 Feb 8;7(1):18. doi: 10.1038/s41522-021-00189-5. NPJ Biofilms Microbiomes. 2021. PMID: 33558519 Free PMC article.
-
Transcriptomic and phenotypic analyses identify coregulated, overlapping regulons among PrfA, CtsR, HrcA, and the alternative sigma factors sigmaB, sigmaC, sigmaH, and sigmaL in Listeria monocytogenes.Appl Environ Microbiol. 2011 Jan;77(1):187-200. doi: 10.1128/AEM.00952-10. Epub 2010 Oct 29. Appl Environ Microbiol. 2011. PMID: 21037293 Free PMC article.
References
-
- Bergmann, B., D. Raffelsbauer, M. Kuhn, M. Goetz, S. Hom, and W. Goebel. 2002. InlA- but not InlB-mediated internalization of Listeria monocytogenes by non-phagocytic mammalian cells needs the support of other internalins. Mol. Microbiol. 43:557-570. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

