Temperature-dependent expression of Listeria monocytogenes internalin and internalin-like genes suggests functional diversity of these proteins among the listeriae
- PMID: 17337561
- PMCID: PMC1892884
- DOI: 10.1128/AEM.02923-06
Temperature-dependent expression of Listeria monocytogenes internalin and internalin-like genes suggests functional diversity of these proteins among the listeriae
Abstract
The Listeria monocytogenes genome contains genes encoding several internalins and internalin-like proteins. As L. monocytogenes is present in many environments and can infect numerous, diverse host species, the environmental temperature was hypothesized to be a signal that might affect internalin gene transcription. A subgenomic microarray was used to investigate temperature-dependent transcription of 24 members of the internalin gene family in L. monocytogenes 10403S. The levels of internalin gene transcripts for cells grown at 37 degrees C were compared to the levels of transcripts for cells grown at 16, 30, and 42 degrees C using competitive microarray hybridization, and the results were confirmed by performing quantitative reverse transcriptase PCR for 14 internalin genes. Based on these studies, the internalin genes can be grouped into the following five temperature-dependent categories: (i) four sigma(B)-dependent internalin genes (inlC2, inlD, lmo0331, and lmo0610) with the highest levels of transcripts at 16 degrees C and generally the lowest levels of transcripts at 37 degrees C; (ii) three partially PrfA-dependent internalin genes (inlA, inlB, and inlC) with the lowest levels of transcripts at 16 degrees C and the highest levels of transcripts at 37 and 42 degrees C; (iii) four genes (inlG, inlJ, lmo0514, and lmo1290) with the lowest levels of transcripts at 16 degrees C and the highest levels of transcripts at 30 and/or 37 degrees C; (iv) one gene (lmo0327) with the highest levels of transcripts at 16 degrees C and low levels of transcripts at higher temperatures; and (v) 12 internalin genes with no differences in the levels of transcripts at the temperatures used in this study. The temperature-dependent transcription patterns suggest that the relative importance of different internalins varies by environment, which may provide insight into the specific functions of these proteins.
Figures

 ). The levels of transcripts were determined by qRT-PCR for L. monocytogenes 10403S grown to the early stationary phase at 16, 30, 37, or 42°C. The error bars indicate one standard deviation based on three independent replicates.
). The levels of transcripts were determined by qRT-PCR for L. monocytogenes 10403S grown to the early stationary phase at 16, 30, 37, or 42°C. The error bars indicate one standard deviation based on three independent replicates.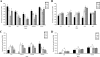
Similar articles
-
Differential regulation of Listeria monocytogenes internalin and internalin-like genes by sigmaB and PrfA as revealed by subgenomic microarray analyses.Foodborne Pathog Dis. 2008 Aug;5(4):417-35. doi: 10.1089/fpd.2008.0085. Foodborne Pathog Dis. 2008. PMID: 18713061 Free PMC article.
-
Contributions of six lineage-specific internalin-like genes to invasion efficiency of Listeria monocytogenes.Foodborne Pathog Dis. 2009 Jan-Feb;6(1):57-70. doi: 10.1089/fpd.2008.0140. Foodborne Pathog Dis. 2009. PMID: 19014275
-
The alternative sigma factor sigma B and the virulence gene regulator PrfA both regulate transcription of Listeria monocytogenes internalins.Appl Environ Microbiol. 2007 May;73(9):2919-30. doi: 10.1128/AEM.02664-06. Epub 2007 Mar 2. Appl Environ Microbiol. 2007. PMID: 17337550 Free PMC article.
-
Role of internalin proteins in the pathogenesis of Listeria monocytogenes.Mol Microbiol. 2021 Dec;116(6):1407-1419. doi: 10.1111/mmi.14836. Epub 2021 Nov 7. Mol Microbiol. 2021. PMID: 34704304 Review.
-
Is the exoproteome important for bacterial pathogenesis? Lessons learned from interstrain exoprotein diversity in Listeria monocytogenes grown at different temperatures.OMICS. 2014 Sep;18(9):553-69. doi: 10.1089/omi.2013.0151. Epub 2014 Aug 15. OMICS. 2014. PMID: 25127015 Review.
Cited by
-
Lmo0171, a novel internalin-like protein, determines cell morphology of Listeria monocytogenes and its ability to invade human cell lines.Curr Microbiol. 2015 Feb;70(2):267-74. doi: 10.1007/s00284-014-0715-4. Epub 2014 Oct 17. Curr Microbiol. 2015. PMID: 25323012 Free PMC article.
-
The Listeria monocytogenes InlC protein interferes with innate immune responses by targeting the I{kappa}B kinase subunit IKK{alpha}.Proc Natl Acad Sci U S A. 2010 Oct 5;107(40):17333-8. doi: 10.1073/pnas.1007765107. Epub 2010 Sep 20. Proc Natl Acad Sci U S A. 2010. PMID: 20855622 Free PMC article.
-
Comparative analysis of the sigma B-dependent stress responses in Listeria monocytogenes and Listeria innocua strains exposed to selected stress conditions.Appl Environ Microbiol. 2008 Jan;74(1):158-71. doi: 10.1128/AEM.00951-07. Epub 2007 Nov 16. Appl Environ Microbiol. 2008. PMID: 18024685 Free PMC article.
-
Microarray-based characterization of the Listeria monocytogenes cold regulon in log- and stationary-phase cells.Appl Environ Microbiol. 2007 Oct;73(20):6484-98. doi: 10.1128/AEM.00897-07. Epub 2007 Aug 24. Appl Environ Microbiol. 2007. PMID: 17720827 Free PMC article.
-
Rhizobacteria Impact Colonization of Listeria monocytogenes on Arabidopsis thaliana Roots.Appl Environ Microbiol. 2021 Nov 10;87(23):e0141121. doi: 10.1128/AEM.01411-21. Epub 2021 Sep 22. Appl Environ Microbiol. 2021. PMID: 34550783 Free PMC article.
References
-
- Bergmann, B., D. Raffelsbauer, M. Kuhn, M. Goetz, S. Hom, and W. Goebel. 2002. InlA- but not InlB-mediated internalization of Listeria monocytogenes by non-phagocytic mammalian cells needs the support of other internalins. Mol. Microbiol. 43:557-570. - PubMed
-
- Bishop, D. K., and D. J. Hinrichs. 1987. Adoptive transfer of immunity to Listeria monocytogenes. The influence of in vitro stimulation on lymphocyte subset requirements. J. Immunol. 139:2005-2009. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

