Characterization of extended-host-range pseudo-T-even bacteriophage Kpp95 isolated on Klebsiella pneumoniae
- PMID: 17337566
- PMCID: PMC1855606
- DOI: 10.1128/AEM.02113-06
Characterization of extended-host-range pseudo-T-even bacteriophage Kpp95 isolated on Klebsiella pneumoniae
Abstract
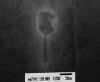
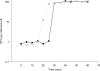
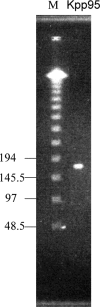
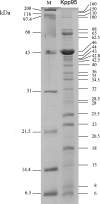
Kpp95, isolated on Klebsiella pneumoniae, is a bacteriophage with the morphology of T4-type phages and is capable of rapid lysis of host cells. Its double-stranded genomic DNA (ca. 175 kb, estimated by pulsed-field gel electrophoresis) can be cut only by restriction endonucleases with a cleavage site flanked either by A and T or by T, as tested, suggesting that it contains the modified derivative(s) of G and/or C. Over 26 protein bands were visualized upon sodium dodecyl sulfate-polyacrylamide gel electrophoresis of the virion proteins. N-terminal sequencing indicated that the most abundant band (46 kDa) is the major coat protein (gp23) which has been cleaved from a signal peptide likely with a length similar to that of T4. Phylogenetic analyses based on the sequences of the central region (263 amino acid residues) of gp23 and the full length of gp18 and gp19 placed Kpp95 among the pseudo-T-even subgroup, most closely related to the coliphage JS98. In addition to being able to lyse many extended-spectrum beta-lactamase strains of K. pneumoniae, Kpp95 can lyse Klebsiella oxytoca, Enterobacter agglomerans, and Serratia marcescens cells. Thus, Kpp95 deserves further studies for development as a component of a therapeutic cocktail, owing to its high efficiencies of host lysis plus extended host range.
Figures
References
-
- Ackermann, H. W., and H. M. Krisch. 1997. A catalogue of T4-type bacteriophages. Arch. Virol. 142:2329-2345. - PubMed
-
- Alisky, J., K. Iczkowski, A. Rapoport, and N. Troitsky. 1998. Bacteriophages show promise as antimicrobial agents. J. Infect. 36:5-15. - PubMed
-
- Benedi, V. J., M. Regue, S. Alberti, S. Camprubi, and J. M. Tomas. 1991. Influence of environmental conditions on infection of Klebsiella pneumoniae by two different types of bacteriophages. Can. J. Microbiol. 37:270-275. - PubMed
-
- Berkner, K. L., and W. R. Folk. 1977. EcoRI cleavage and methylation of DNAs containing modified pyrimidines in the recognition sequence. J. Biol. Chem. 252:3185-3193. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

