Elongation factor Tu3 (EF-Tu3) from the kirromycin producer Streptomyces ramocissimus Is resistant to three classes of EF-Tu-specific inhibitors
- PMID: 17337575
- PMCID: PMC1855904
- DOI: 10.1128/JB.01810-06
Elongation factor Tu3 (EF-Tu3) from the kirromycin producer Streptomyces ramocissimus Is resistant to three classes of EF-Tu-specific inhibitors
Abstract
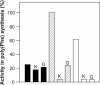
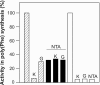
The antibiotic kirromycin inhibits prokaryotic protein synthesis by immobilizing elongation factor Tu (EF-Tu) on the elongating ribosome. Streptomyces ramocissimus, the producer of kirromycin, contains three tuf genes. While tuf1 and tuf2 encode kirromycin-sensitive EF-Tu species, the function of tuf3 is unknown. Here we demonstrate that EF-Tu3, in contrast to EF-Tu1 and EF-Tu2, is resistant to three classes of EF-Tu-targeted antibiotics: kirromycin, pulvomycin, and GE2270A. A mixture of EF-Tu1 and EF-Tu3 was sensitive to kirromycin and resistant to GE2270A, in agreement with the described modes of action of these antibiotics. Transcription of tuf3 was observed during exponential growth and ceased upon entry into stationary phase and therefore did not correlate with the appearance of kirromycin in stationary phase; thus, it is unlikely that EF-Tu3 functions as a resistant alternative for EF-Tu1. EF-Tu3 from Streptomyces coelicolor A3(2) was also resistant to kirromycin and GE2270A, suggesting that multiple antibiotic resistance is an intrinsic feature of EF-Tu3 species. The GE2270A-resistant character of EF-Tu3 demonstrated that this divergent elongation factor is capable of substituting for EF-Tu1 in vivo.
Figures





Similar articles
-
The unique tuf2 gene from the kirromycin producer Streptomyces ramocissimus encodes a minor and kirromycin-sensitive elongation factor Tu.J Bacteriol. 2002 Aug;184(15):4211-8. doi: 10.1128/JB.184.15.4211-4218.2002. J Bacteriol. 2002. PMID: 12107139 Free PMC article.
-
Elongation factor Tu1 of the antibiotic GE2270A producer Planobispora rosea has an unexpected resistance profile against EF-Tu targeted antibiotics.Biochem Biophys Res Commun. 1997 Jan 13;230(2):320-6. doi: 10.1006/bbrc.1996.5947. Biochem Biophys Res Commun. 1997. PMID: 9016775
-
Three tuf-like genes in the kirromycin producer Streptomyces ramocissimus.Microbiology (Reading). 1994 Apr;140 ( Pt 4):983-98. doi: 10.1099/00221287-140-4-983. Microbiology (Reading). 1994. PMID: 8012612
-
Inhibitory mechanisms of antibiotics targeting elongation factor Tu.Curr Protein Pept Sci. 2002 Feb;3(1):121-31. doi: 10.2174/1389203023380855. Curr Protein Pept Sci. 2002. PMID: 12370016 Review.
-
Antibiotic resistance mechanisms of mutant EF-Tu species in Escherichia coli.Biochem Cell Biol. 1995 Nov-Dec;73(11-12):1167-77. doi: 10.1139/o95-126. Biochem Cell Biol. 1995. PMID: 8722034 Review.
Cited by
-
Comparison of Antibiotic Resistance Mechanisms in Antibiotic-Producing and Pathogenic Bacteria.Molecules. 2019 Sep 21;24(19):3430. doi: 10.3390/molecules24193430. Molecules. 2019. PMID: 31546630 Free PMC article. Review.
-
Comparison of Soil Microbial Composition in Rhizospheres Between Wilt Disease-Resistant and Susceptible Melon Varieties.Microorganisms. 2025 Feb 18;13(2):444. doi: 10.3390/microorganisms13020444. Microorganisms. 2025. PMID: 40005810 Free PMC article.
-
Engineering Streptomyces coelicolor for heterologous expression of the thiopeptide GE2270A-A cautionary tale.J Ind Microbiol Biotechnol. 2024 Dec 31;52:kuaf019. doi: 10.1093/jimb/kuaf019. J Ind Microbiol Biotechnol. 2024. PMID: 40679465 Free PMC article.
-
Identification and activation of novel biosynthetic gene clusters by genome mining in the kirromycin producer Streptomyces collinus Tü 365.J Ind Microbiol Biotechnol. 2016 Mar;43(2-3):277-91. doi: 10.1007/s10295-015-1685-7. Epub 2015 Oct 3. J Ind Microbiol Biotechnol. 2016. PMID: 26433383
-
Polyketide Bioderivatization Using the Promiscuous Acyltransferase KirCII.ACS Synth Biol. 2017 Mar 17;6(3):421-427. doi: 10.1021/acssynbio.6b00341. Epub 2017 Feb 22. ACS Synth Biol. 2017. PMID: 28206741 Free PMC article.
References
-
- Abdulkarim, F., L. Liljas, and D. Hughes. 1994. Mutations to kirromycin resistance occur in the interface of domains I and III of EF-Tu.GTP. FEBS Lett. 352:118-122. - PubMed
-
- Abel, K., M. D. Yoder, R. Hilgenfeld, and F. Jurnak. 1996. An α to β conformational switch in EF-Tu. Structure 4:1153-1159. - PubMed
-
- Berchtold, H., L. Reshetnikova, C. O. A. Reiser, N. K. Schirmer, M. Sprinzl, and R. Hilgenfeld. 1993. Crystal structure of active elongation factor Tu reveals major domain rearrangements. Nature 365:126-132. - PubMed
-
- Boon, K., I. Krab, A. Parmeggiani, L. Bosch, and B. Kraal. 1995. Substitution of Arg230 and Arg233 in Escherichia coli elongation factor Tu strongly enhances its pulvomycin resistance. Eur. J. Biochem. 227:816-822. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

