A putative gene cluster for aminoarabinose biosynthesis is essential for Burkholderia cenocepacia viability
- PMID: 17337576
- PMCID: PMC1855895
- DOI: 10.1128/JB.00153-07
A putative gene cluster for aminoarabinose biosynthesis is essential for Burkholderia cenocepacia viability
Abstract
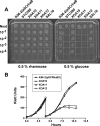
Using a conditional mutagenesis strategy we demonstrate here that a gene cluster encoding putative aminoarabinose (Ara4N) biosynthesis enzymes is essential for the viability of Burkholderia cenocepacia. Loss of viability is associated with dramatic changes in bacterial cell morphology and ultrastructure, increased permeability to propidium iodide, and sensitivity to sodium dodecyl sulfate, suggesting a general cell envelope defect caused by the lack of Ara4N.
Figures



References
-
- Bader, M. W., S. Sanowar, M. E. Daley, A. R. Schneider, U. Cho, W. Xu, R. E. Klevit, H. Le Moual, and S. I. Miller. 2005. Recognition of antimicrobial peptides by a bacterial sensor kinase. Cell 122:461-472. - PubMed
-
- Bailey, A. M., S. Mahapatra, P. J. Brennan, and D. C. Crick. 2002. Identification, cloning, purification, and enzymatic characterization of Mycobacterium tuberculosis 1-deoxy-d-xylulose 5-phosphate synthase. Glycobiology 12:813-820. - PubMed
-
- Breazeale, S. D., A. A. Ribeiro, and C. R. Raetz. 2002. Oxidative decarboxylation of UDP-glucuronic acid in extracts of polymyxin-resistant Escherichia coli. Origin of lipid a species modified with 4-amino-4-deoxy-l-arabinose. J. Biol. Chem. 277:2886-2896. - PubMed
-
- Breazeale, S. D., A. A. Ribeiro, and C. R. H. Raetz. 2003. Origin of lipid A species modified with 4-amino-4-deoxy-l-arabinose in polymyxin-resistant mutants of Escherichia coli: an aminotransferase (arnB) that generates UDP-4-amino-4-deoxy-l-arabinose. J. Biol. Chem. 278:24731-24739. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

