HpdR is a transcriptional activator of Sinorhizobium meliloti hpdA, which encodes a herbicide-targeted 4-hydroxyphenylpyruvate dioxygenase
- PMID: 17337579
- PMCID: PMC1855912
- DOI: 10.1128/JB.01662-06
HpdR is a transcriptional activator of Sinorhizobium meliloti hpdA, which encodes a herbicide-targeted 4-hydroxyphenylpyruvate dioxygenase
Abstract
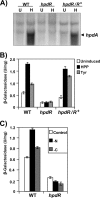
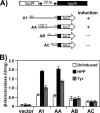
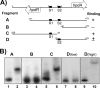
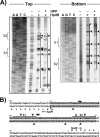
Sinorhizobium meliloti hpdA, which encodes the herbicide target 4-hydroxyphenylpyruvate dioxygenase, is positively regulated by HpdR. Gel mobility shift and DNase I footprinting analyses revealed that HpdR binds to a region that spans two conserved direct-repeat sequences within the hpdR-hpdA intergenic space. HpdR-dependent hpdA transcription occurs in the presence of 4-hydroxyphenylpyruvate, tyrosine, and phenylalanine, as well as during starvation.
Figures

Similar articles
-
The tyrosine degradation gene hppD is transcriptionally activated by HpdA and repressed by HpdR in Streptomyces coelicolor, while hpdA is negatively autoregulated and repressed by HpdR.Mol Microbiol. 2007 Aug;65(4):1064-77. doi: 10.1111/j.1365-2958.2007.05848.x. Epub 2007 Jul 19. Mol Microbiol. 2007. PMID: 17640269
-
Rem, a new transcriptional activator of motility and chemotaxis in Sinorhizobium meliloti.J Bacteriol. 2006 Oct;188(19):6932-42. doi: 10.1128/JB.01902-05. J Bacteriol. 2006. PMID: 16980496 Free PMC article.
-
The upstream region of the nodD3 gene of Sinorhizobium meliloti carries enhancer sequences for the transcriptional activator NtrC.FEMS Microbiol Lett. 1999 Oct 15;179(2):491-9. doi: 10.1111/j.1574-6968.1999.tb08768.x. FEMS Microbiol Lett. 1999. PMID: 10518756
-
Negative Regulation of Ectoine Uptake and Catabolism in Sinorhizobium meliloti: Characterization of the EhuR Gene.J Bacteriol. 2016 Dec 13;199(1):e00119-16. doi: 10.1128/JB.00119-16. Print 2017 Jan 1. J Bacteriol. 2016. PMID: 27795315 Free PMC article.
-
Identification of direct transcriptional target genes of ExoS/ChvI two-component signaling in Sinorhizobium meliloti.J Bacteriol. 2009 Nov;191(22):6833-42. doi: 10.1128/JB.00734-09. Epub 2009 Sep 11. J Bacteriol. 2009. PMID: 19749054 Free PMC article.
Cited by
-
The hdhA gene encodes a haloacid dehalogenase that is regulated by the LysR-type regulator, HdhR, in Sinorhizobium meliloti.Mol Biotechnol. 2013 Jun;54(2):148-57. doi: 10.1007/s12033-012-9556-1. Mol Biotechnol. 2013. PMID: 22638965
-
Comparative transcriptomic analysis reveals the significant pleiotropic regulatory effects of LmbU on lincomycin biosynthesis.Microb Cell Fact. 2020 Feb 12;19(1):30. doi: 10.1186/s12934-020-01298-0. Microb Cell Fact. 2020. PMID: 32050973 Free PMC article.
References
-
- Alexeyev, M. F. 1999. The pKNOCK series of broad-host-range mobilizable suicide vectors for gene knockout and targeted DNA insertion into the chromosome of gram-negative bacteria. BioTechniques. 26:824-828. - PubMed
-
- Barran, L. R., E. S. Bromfield, and D. C. Brown. 2002. Identification and cloning of the bacterial nodulation specificity gene in the Sinorhizobium meliloti-Medicago laciniata symbiosis. Can. J. Microbiol. 48:765-771. - PubMed
-
- Brinkman, A. B., T. J. Ettema, W. M. de Vos, and J. van der Oost. 2003. The Lrp family of transcriptional regulators. Mol. Microbiol. 48:287-294. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

