Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development
- PMID: 17337630
- PMCID: PMC1867377
- DOI: 10.1105/tpc.106.047688
Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development
Abstract
The transcription factor LONG HYPOCOTYL5 (HY5) acts downstream of multiple families of the photoreceptors and promotes photomorphogenesis. Although it is well accepted that HY5 acts to regulate target gene expression, in vivo binding of HY5 to any of its target gene promoters has yet to be demonstrated. Here, we used a chromatin immunoprecipitation procedure to verify suspected in vivo HY5 binding sites. We demonstrated that in vivo association of HY5 with promoter targets is not altered under distinct light qualities or during light-to-dark transition. Coupled with DNA chip hybridization using a high-density 60-nucleotide oligomer microarray that contains one probe for every 500 nucleotides over the entire Arabidopsis thaliana genome, we mapped genome-wide in vivo HY5 binding sites. This analysis showed that HY5 binds preferentially to promoter regions in vivo and revealed >3000 chromosomal sites as putative HY5 binding targets. HY5 binding targets tend to be enriched in the early light-responsive genes and transcription factor genes. Our data thus support a model in which HY5 is a high hierarchical regulator of the transcriptional cascades for photomorphogenesis.
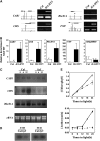
Figures




References
-
- Ang, L.H., Chattopadhyay, S., Wei, N., Oyama, T., Okada, K., Batschauer, A., and Deng, X.W. (1998). Molecular interaction between COP1 and HY5 defines a regulatory switch for light control of Arabidopsis development. Mol. Cell 1 213–222. - PubMed
-
- Arabidopsis Genome Initiative (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408 796–815. - PubMed
-
- Benjamini, Y., and Hochberg, Y. (1995). Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57 289–300.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

