The FLOWERING LOCUS T-like gene family in barley (Hordeum vulgare)
- PMID: 17339225
- PMCID: PMC1893030
- DOI: 10.1534/genetics.106.069500
The FLOWERING LOCUS T-like gene family in barley (Hordeum vulgare)
Abstract
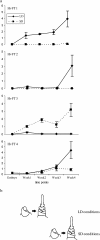
The FLOWERING LOCUS T (FT) gene plays a central role in integrating flowering signals in Arabidopsis because its expression is regulated antagonistically by the photoperiod and vernalization pathways. FT belongs to a family of six genes characterized by a phosphatidylethanolamine-binding protein (PEBP) domain. In rice (Oryza sativa), 19 PEBP genes were previously described, 13 of which are FT-like genes. Five FT-like genes were found in barley (Hordeum vulgare). HvFT1, HvFT2, HvFT3, and HvFT4 were highly homologous to OsFTL2 (the Hd3a QTL), OsFTL1, OsFTL10, and OsFTL12, respectively, and this relationship was supported by comparative mapping. No rice equivalent was found for HvFT5. HvFT1 was highly expressed under long-day (inductive) conditions at the time of the morphological switch of the shoot apex from vegetative to reproductive growth. HvFT2 and HvFT4 were expressed later in development. HvFT1 was therefore identified as the main barley FT-like gene involved in the switch to flowering. Mapping of HvFT genes suggests that they provide important sources of flowering-time variation in barley. HvFTI was a candidate for VRN-H3, a dominant mutation giving precocious flowering, while HvFT3 was a candidate for Ppd-H2, a major QTL affecting flowering time in short days.
Figures


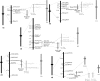
References
-
- Abe, M., Y. Kobayashi, S. Yamamoto, Y. Daimon, A. Yamaguchi et al., 2005. FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309: 1052–1056. - PubMed
-
- An, H. L., C. Roussot, P. Suarez-Lopez, L. Corbesler, C. Vincent et al., 2004. CONSTANS acts in the phloem to regulate a systemic signal that induces photoperiodic flowering of Arabidopsis. Development 131: 3615–3626. - PubMed
-
- Armstead, I. P., L. B. Turner, I. P. King, A. J. Cairns and M. O. Humphreys, 2002. Comparison and integration of genetic maps generated from F-2 and BC1-type mapping populations in perennial ryegrass. Plant Breed. 121: 501–507.
-
- Bäurle, I., and C. Dean, 2006. The timing of developmental transitions in plants. Cell 125: 655–664. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

