mRNAs encoding polarity and exocytosis factors are cotransported with the cortical endoplasmic reticulum to the incipient bud in Saccharomyces cerevisiae
- PMID: 17339339
- PMCID: PMC1899969
- DOI: 10.1128/MCB.01643-06
mRNAs encoding polarity and exocytosis factors are cotransported with the cortical endoplasmic reticulum to the incipient bud in Saccharomyces cerevisiae
Abstract
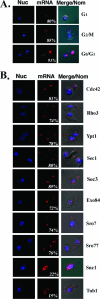
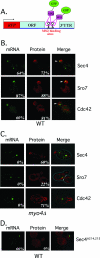
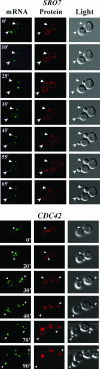
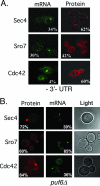
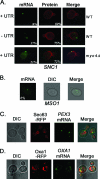
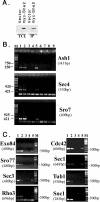
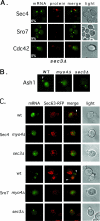
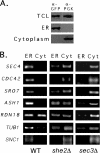
Polarized growth in the budding yeast Saccharomyces cerevisiae depends upon the asymmetric localization and enrichment of polarity and secretion factors at the membrane prior to budding. We examined how these factors (i.e., Cdc42, Sec4, and Sro7) reach the bud site and found that their respective mRNAs localize to the tip of the incipient bud prior to nuclear division. Asymmetric mRNA localization depends upon factors that facilitate ASH1 mRNA localization (e.g., the 3' untranslated region, She proteins 1 to 5, Puf6, actin cytoskeleton, and a physical association with She2). mRNA placement precedes protein enrichment and subsequent bud emergence, implying that mRNA localization contributes to polarization. Correspondingly, mRNAs encoding proteins which are not asymmetrically distributed (i.e., Snc1, Mso1, Tub1, Pex3, and Oxa1) are not polarized. Finally, mutations which affect cortical endoplasmic reticulum (ER) entry and anchoring in the bud (myo4Delta, sec3Delta, and srp101) also affect asymmetric mRNA localization. Bud-localized mRNAs, including ASH1, were found to cofractionate with ER microsomes in a She2- and Sec3-dependent manner; thus, asymmetric mRNA transport and cortical ER inheritance are connected processes in yeast.
Figures

References
-
- Aronov, S., and J. E. Gerst. 2004. Involvement of the late secretory pathway in actin regulation and mRNA transport in yeast. J. Biol. Chem. 279:36962-36971. - PubMed
-
- Barral, Y., V. Mermall, M. S. Mooseker, and M. Snyder. 2000. Compartmentalization of the cell cortex by septins is required for maintenance of cell polarity in yeast. Mol. Cell 5:841-851. - PubMed
-
- Baum, B. 2004. Animal development: crowd control. Curr. Biol. 14:R716—R718. - PubMed
-
- Beach, D. L., E. D. Salmon, and K. Bloom. 1999. Localization and anchoring of mRNA in budding yeast. Curr. Biol. 9:569-578. - PubMed
-
- Bertrand, E., P. Chartrand, M. Schaefer, S. M. Shenoy, R. H. Singer, and R. M. Long. 1998. Localization of ASH1 mRNA particles in living yeast. Mol. Cell 2:437-445. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
