Function of a conserved checkpoint recruitment domain in ATRIP proteins
- PMID: 17339343
- PMCID: PMC1899971
- DOI: 10.1128/MCB.02238-06
Function of a conserved checkpoint recruitment domain in ATRIP proteins
Abstract
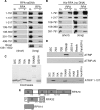
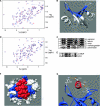
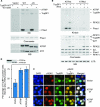
The ATR (ATM and Rad3-related) kinase is essential to maintain genomic integrity. ATR is recruited to DNA lesions in part through its association with ATR-interacting protein (ATRIP), which in turn interacts with the single-stranded DNA binding protein RPA (replication protein A). In this study, a conserved checkpoint protein recruitment domain (CRD) in ATRIP orthologs was identified by biochemical mapping of the RPA binding site in combination with nuclear magnetic resonance, mutagenesis, and computational modeling. Mutations in the CRD of the Saccharomyces cerevisiae ATRIP ortholog Ddc2 disrupt the Ddc2-RPA interaction, prevent proper localization of Ddc2 to DNA breaks, sensitize yeast to DNA-damaging agents, and partially compromise checkpoint signaling. These data demonstrate that the CRD is critical for localization and optimal DNA damage responses. However, the stimulation of ATR kinase activity by binding of topoisomerase binding protein 1 (TopBP1) to ATRIP-ATR can occur independently of the interaction of ATRIP with RPA. Our results support the idea of a multistep model for ATR activation that requires separable localization and activation functions of ATRIP.
Figures



References
-
- Amberg, D. C., D. J. Burke, and J. E. Strathern. 2005. Methods in yeast genetics: a Cold Spring Harbor laboratory course manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
-
- Bermudez, V. P., L. A. Lindsey-Boltz, A. J. Cesare, Y. Maniwa, J. D. Griffith, J. Hurwitz, and A. Sancar. 2003. Loading of the human 9-1-1 checkpoint complex onto DNA by the checkpoint clamp loader hRad17-replication factor C complex in vitro. Proc. Natl. Acad. Sci. USA 100:1633-1638. - PMC - PubMed
-
- Binz, S. K., Y. Lao, D. F. Lowry, and M. S. Wold. 2003. The phosphorylation domain of the 32-kDa subunit of replication protein A (RPA) modulates RPA-DNA interactions. Evidence for an intersubunit interaction. J. Biol. Chem. 278:35584-35591. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
