The evolutionary history of human DNA transposons: evidence for intense activity in the primate lineage
- PMID: 17339369
- PMCID: PMC1832089
- DOI: 10.1101/gr.5826307
The evolutionary history of human DNA transposons: evidence for intense activity in the primate lineage
Abstract
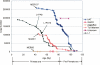
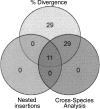
Class 2, or DNA transposons, make up approximately 3% of the human genome, yet the evolutionary history of these elements has been largely overlooked and remains poorly understood. Here we carried out the first comprehensive analysis of the activity of human DNA transposons over the course of primate evolution using three independent computational methods. First, we conducted an exhaustive search for human DNA transposons nested within L1 and Alu elements known to be primate specific. Second, we assessed the presence/absence of 794 human DNA transposons at orthologous positions in 10 mammalian species using sequence data generated by The ENCODE Project. These two approaches, which do not rely upon sequence divergence, allowed us to classify DNA transposons into three different categories: anthropoid specific (40-63 My), primate specific (64-80 My), and eutherian wide (81-150 My). Finally, we used this data to calculate the substitution rates of DNA transposons for each category and refine the age of each family based on the average percent divergence of individual copies to their consensus. Based on these combined methods, we can confidently estimate that at least 40 human DNA transposon families, representing approximately 98,000 elements ( approximately 33 Mb) in the human genome, have been active in the primate lineage. There was a cessation in the transpositional activity of DNA transposons during the later phase of the primate radiation, with no evidence of elements younger than approximately 37 My. This data points to intense activity of DNA transposons during the mammalian radiation and early primate evolution, followed, apparently, by their mass extinction in an anthropoid primate ancestor.
Figures

References
-
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J., Gish W., Miller W., Myers E.W., Lipman D.J., Miller W., Myers E.W., Lipman D.J., Myers E.W., Lipman D.J., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. - PubMed
-
- Auge-Gouillou C., Bigot Y., Pollet N., Hamelin M.H., Meunier-Rotival M., Periquet G., Bigot Y., Pollet N., Hamelin M.H., Meunier-Rotival M., Periquet G., Pollet N., Hamelin M.H., Meunier-Rotival M., Periquet G., Hamelin M.H., Meunier-Rotival M., Periquet G., Meunier-Rotival M., Periquet G., Periquet G. Human and other mammalian genomes contain transposons of the mariner family. FEBS Lett. 1995;368:541–546. - PubMed
-
- Caceres M., Ranz J.M., Barbadilla A., Long M., Ruiz A., Ranz J.M., Barbadilla A., Long M., Ruiz A., Barbadilla A., Long M., Ruiz A., Long M., Ruiz A., Ruiz A. Generation of a widespread Drosophila inversion by a transposable element. Science. 1999;285:415–418. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
