Propagation of centromeric chromatin requires exit from mitosis
- PMID: 17339380
- PMCID: PMC2064054
- DOI: 10.1083/jcb.200701066
Propagation of centromeric chromatin requires exit from mitosis
Abstract
Centromeres direct chromosomal inheritance by nucleating assembly of the kinetochore, a large multiprotein complex required for microtubule attachment during mitosis. Centromere identity in humans is epigenetically determined, with no DNA sequence either necessary or sufficient. A prime candidate for the epigenetic mark is assembly into centromeric chromatin of centromere protein A (CENP-A), a histone H3 variant found only at functional centromeres. A new covalent fluorescent pulse-chase labeling approach using SNAP tagging has now been developed and is used to demonstrate that CENP-A bound to a mature centromere is quantitatively and equally partitioned to sister centromeres generated during S phase, thereby remaining stably associated through multiple cell divisions. Loading of nascent CENP-A on the megabase domains of replicated centromere DNA is shown to require passage through mitosis but not microtubule attachment. Very surprisingly, assembly and stabilization of new CENP-A-containing nucleosomes is restricted exclusively to the subsequent G1 phase, demonstrating direct coupling between progression through mitosis and assembly/maturation of the next generation of centromeres.
Figures
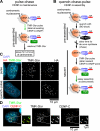
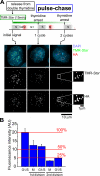


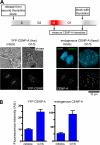
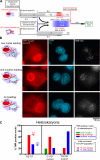
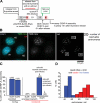
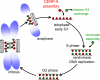
Comment in
-
Centromeric chromatin gets loaded.J Cell Biol. 2007 Mar 12;176(6):735-6. doi: 10.1083/jcb.200702020. Epub 2007 Mar 5. J Cell Biol. 2007. PMID: 17339381 Free PMC article.
References
-
- Amor, D.J., P. Kalitsis, H. Sumer, and K.H. Choo. 2004. b. Building the centromere: from foundation proteins to 3D organization. Trends Cell Biol. 14:359–368. - PubMed
-
- Black, B.E., D.R. Foltz, S. Chakravarthy, K. Luger, V.L. Woods Jr., and D.W. Cleveland. 2004. Structural determinants for generating centromeric chromatin. Nature. 430:578–582. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

