The pharmacogenetics research network: from SNP discovery to clinical drug response
- PMID: 17339863
- PMCID: PMC5006950
- DOI: 10.1038/sj.clpt.6100087
The pharmacogenetics research network: from SNP discovery to clinical drug response
Abstract
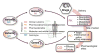
The NIH Pharmacogenetics Research Network (PGRN) is a collaborative group of investigators with a wide range of research interests, but all attempting to correlate drug response with genetic variation. Several research groups concentrate on drugs used to treat specific medical disorders (asthma, depression, cardiovascular disease, addiction of nicotine, and cancer), whereas others are focused on specific groups of proteins that interact with drugs (membrane transporters and phase II drug-metabolizing enzymes). The diverse scientific information is stored and annotated in a publicly accessible knowledge base, the Pharmacogenetics and Pharmacogenomics Knowledge base (PharmGKB). This report highlights selected achievements and scientific approaches as well as hypotheses about future directions of each of the groups within the PGRN. Seven major topics are included: informatics (PharmGKB), cardiovascular, pulmonary, addiction, cancer, transport, and metabolism.
Conflict of interest statement
D.M. Roden (GlaxoSmithKline, Pfizer, Inc., AstraZeneca, Abbott Laboratories, Novartis, 1st Genetic Trust), R.M. Krauss (Abbott Laboratories, AstraZeneca, Bristol-Myers Squibb, Merck & Co. Inc., Pfizer Inc., International Dairy Foods Association), M.J. Ratain (Prometheus, Genzyme Corp., Genentech); S.T. Weiss (Glaxo-Wellcome, Roche Pharmaceuticals, Millennium Pharmaceuticals, Genentech, Shering-Plough, Variagenics, Genome Therapeutics, Merck Frost). Stock ownership or operations (other than mutual funds): M.J. Ratain (Variagenics, Nuvelo, Applera). Grants received: D.M. Roden (1st Genetic Trusty), R.M. Krauss (King, Merck Schering Plough, Pfizer Inc.), S.T. Weiss (Glaxo-Wellcome, AstraZeneca, Pfizer). Patents received: M.J. Ratain (National Institutes of Health), M.V. Relling (National Institutes of Health).
Figures



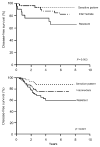
References
-
- Klein TE, et al. Integrating genotype and phenotype information: an overview of the PharmGKB project. Pharmacogenomics J. 2001;1:167–170. - PubMed
-
- Fields LE, et al. The burden of adult hypertension in the United States 1999–2000: a rising tide. Hypertension. 2004;44:398–404. - PubMed
-
- Chobanian AV, et al. The seventh report of the Joint National Committee on Prevention, Detection, Evaluation, and Treatment of High Blood Pressure: The JNC 7 Report. JAMA. 2003;289:2560–2571. - PubMed
-
- Materson BJ, et al. Single-drug therapy for hypertension in men. A comparison of six antihypertensive agents with placebo. The Department of Veterans Affairs Cooperative Study Group on Antihypertensive Agents. N Engl J Med. 1993;328:914–921. [see comments] [published erratum appears in N Engl J Med 330, 1689 (1994)] - PubMed
-
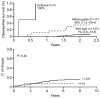
- Turner ST, Chapman AB, Schwartz GL, Boerwinkle E. Effects of endothelial nitric oxide synthase, alpha-adducin, and other candidate gene polymorphisms on blood pressure response to hydrochlorothiazide. Am J Hypertens. 2003;16:834–839. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- U19 GM061388/GM/NIGMS NIH HHS/United States
- U01 GM063340/GM/NIGMS NIH HHS/United States
- U19 HL065962/HL/NHLBI NIH HHS/United States
- U01 HL065899/HL/NHLBI NIH HHS/United States
- U01 HL065962/HL/NHLBI NIH HHS/United States
- U01 GM061388/GM/NIGMS NIH HHS/United States
- GM61374/GM/NIGMS NIH HHS/United States
- GM61390/GM/NIGMS NIH HHS/United States
- U19 GM061390/GM/NIGMS NIH HHS/United States
- U19 HL069757/HL/NHLBI NIH HHS/United States
- GM61737/GM/NIGMS NIH HHS/United States
- R24 GM061374/GM/NIGMS NIH HHS/United States
- HL65899/HL/NHLBI NIH HHS/United States
- HL65962/HL/NHLBI NIH HHS/United States
- U01 GM061393/GM/NIGMS NIH HHS/United States
- GM61388/GM/NIGMS NIH HHS/United States
- U01 GM074492/GM/NIGMS NIH HHS/United States
- GM61393/GM/NIGMS NIH HHS/United States
- HL60757/HL/NHLBI NIH HHS/United States
- GM74492/GM/NIGMS NIH HHS/United States
- GM63340/GM/NIGMS NIH HHS/United States
- U01 GM061390/GM/NIGMS NIH HHS/United States
- U01 GM061374/GM/NIGMS NIH HHS/United States
- GM74518/GM/NIGMS NIH HHS/United States
- U01 GM061373/GM/NIGMS NIH HHS/United States
- WT_/Wellcome Trust/United Kingdom
- U01 HL069757/HL/NHLBI NIH HHS/United States
- U01 GM074518/GM/NIGMS NIH HHS/United States
- DA20830/DA/NIDA NIH HHS/United States
- U01 DA020830/DA/NIDA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources

