Tat competes with HEXIM1 to increase the active pool of P-TEFb for HIV-1 transcription
- PMID: 17341462
- PMCID: PMC1874611
- DOI: 10.1093/nar/gkm063
Tat competes with HEXIM1 to increase the active pool of P-TEFb for HIV-1 transcription
Abstract
Human immunodeficiency virus type 1 (HIV-1) transcriptional transactivator (Tat) recruits the positive transcription elongation factor b (P-TEFb) to the viral promoter. Consisting of cyclin dependent kinase 9 (Cdk9) and cyclin T1, P-TEFb phosphorylates RNA polymerase II and the negative transcription elongation factor to stimulate the elongation of HIV-1 genes. A major fraction of nuclear P-TEFb is sequestered into a transcriptionally inactive 7SK small nuclear ribonucleoprotein (snRNP) by the coordinated actions of the 7SK small nuclear RNA (snRNA) and hexamethylene bisacetamide (HMBA) induced protein 1 (HEXIM1). In this study, we demonstrate that Tat prevents the formation of and also releases P-TEFb from the 7SK snRNP in vitro and in vivo. This ability of Tat depends on the integrity of its N-terminal activation domain and stems from the high affinity interaction between Tat and cyclin T1, which allows Tat to directly displace HEXIM1 from cyclin T1. Furthermore, we find that in contrast to the Tat-independent activation of the HIV-1 promoter, Tat-dependent HIV-1 transcription is largely insensitive to the inhibition by HEXIM1. Finally, primary blood lymphocytes display a reduced amount of the endogenous 7SK snRNP upon HIV-1 infection. All these data are consistent with the model that Tat not only recruits but also increases the active pool of P-TEFb for efficient HIV-1 transcription.
Figures
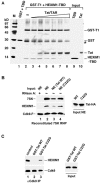



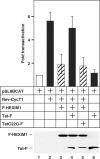
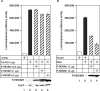

References
-
- Yik JH, Chen R, Nishimura R, Jennings JL, Link AJ, Zhou Q. Inhibition of P-TEFb (CDK9/Cyclin T) kinase and RNA polymerase II transcription by the coordinated actions of HEXIM1 and 7SK snRNA. Mol Cell. 2003;12:971–982. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

