Does the DNA barcoding gap exist? - a case study in blue butterflies (Lepidoptera: Lycaenidae)
- PMID: 17343734
- PMCID: PMC1838910
- DOI: 10.1186/1742-9994-4-8
Does the DNA barcoding gap exist? - a case study in blue butterflies (Lepidoptera: Lycaenidae)
Abstract
Background: DNA barcoding, i.e. the use of a 648 bp section of the mitochondrial gene cytochrome c oxidase I, has recently been promoted as useful for the rapid identification and discovery of species. Its success is dependent either on the strength of the claim that interspecific variation exceeds intraspecific variation by one order of magnitude, thus establishing a "barcoding gap", or on the reciprocal monophyly of species.
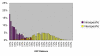
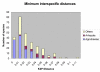
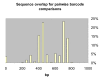
Results: We present an analysis of intra- and interspecific variation in the butterfly family Lycaenidae which includes a well-sampled clade (genus Agrodiaetus) with a peculiar characteristic: most of its members are karyologically differentiated from each other which facilitates the recognition of species as reproductively isolated units even in allopatric populations. The analysis shows that there is an 18% overlap in the range of intra- and interspecific COI sequence divergence due to low interspecific divergence between many closely related species. In a Neighbour-Joining tree profile approach which does not depend on a barcoding gap, but on comprehensive sampling of taxa and the reciprocal monophyly of species, at least 16% of specimens with conspecific sequences in the profile were misidentified. This is due to paraphyly or polyphyly of conspecific DNA sequences probably caused by incomplete lineage sorting.
Conclusion: Our results indicate that the "barcoding gap" is an artifact of insufficient sampling across taxa. Although DNA barcodes can help to identify and distinguish species, we advocate using them in combination with other data, since otherwise there would be a high probability that sequences are misidentified. Although high differences in DNA sequences can help to identify cryptic species, a high percentage of well-differentiated species has similar or even identical COI sequences and would be overlooked in an isolated DNA barcoding approach.
Figures



References
-
- Hebert PD, Ratnasingham S, deWaard JR. Barcoding animal life: cytochrome c oxidase subunit 1 divergences among closely related species. Proc Biol Sci. 2003;270 Suppl 1:S96–9. doi: 10.1098/rsbl.2003.0025. http://www.journals.royalsoc.ac.uk/openurl.asp?genre=article&id=doi:10.1... - DOI - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

