Analysis of read length limiting factors in Pyrosequencing chemistry
- PMID: 17343818
- PMCID: PMC1978072
- DOI: 10.1016/j.ab.2007.02.002
Analysis of read length limiting factors in Pyrosequencing chemistry
Abstract
Pyrosequencing is a bioluminometric DNA sequencing technique that measures the release of pyrophosphate during DNA synthesis. The amount of pyrophosphate is proportionally converted into visible light by a cascade of enzymatic reactions. Pyrosequencing has heretofore been used for generating short sequence reads (1-100 nucleotides) because certain factors limit the system's ability to perform longer reads accurately. In this study, we have characterized the main read length limiting factors in both three-enzyme and four-enzyme Pyrosequencing systems. A new simulation model was developed to simulate the read length of both systems based on the inhibitory factors in the chemical equations governing each enzymatic cascade. Our results indicate that nonsynchronized extension limits the obtained read length, albeit to a different extent for each system. In the four-enzyme system, nonsynchronized extension due mainly to a decrease in apyrase's efficiency in degrading excess nucleotides proves to be the main limiting factor of read length. Replacing apyrase with a washing step for removal of excess nucleotide proves to be essential in improving the read length of Pyrosequencing. The main limiting factor of the three-enzyme system is shown to be loss of DNA fragments during the washing step. If this loss is minimized to 0.1% per washing cycle, the read length of Pyrosequencing would be well beyond 300 bases.
Figures






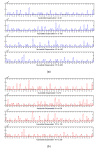
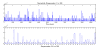
Similar articles
-
A multi-enzyme model for Pyrosequencing.Nucleic Acids Res. 2004 Dec 2;32(21):e166. doi: 10.1093/nar/gnh159. Nucleic Acids Res. 2004. PMID: 15576673 Free PMC article.
-
Pyrosequencing: history, biochemistry and future.Clin Chim Acta. 2006 Jan;363(1-2):83-94. doi: 10.1016/j.cccn.2005.04.038. Epub 2005 Sep 13. Clin Chim Acta. 2006. PMID: 16165119 Review.
-
[Development of 3-enzyme pyrosequencing system and its application in rapid diagnosis of Down's syndrome].Yi Chuan. 2010 May;32(5):517-23. doi: 10.3724/sp.j.1005.2010.00517. Yi Chuan. 2010. PMID: 20466643 Chinese.
-
A sequencing method based on real-time pyrophosphate.Science. 1998 Jul 17;281(5375):363, 365. doi: 10.1126/science.281.5375.363. Science. 1998. PMID: 9705713 No abstract available.
-
Pyrosequencing: nucleotide sequencing technology with bacterial genotyping applications.Expert Rev Mol Diagn. 2005 Nov;5(6):947-53. doi: 10.1586/14737159.5.6.947. Expert Rev Mol Diagn. 2005. PMID: 16255635 Review.
Cited by
-
High-throughput homogeneous mass cleave assay technology for the diagnosis of autosomal recessive Parkinson's disease.J Mol Diagn. 2008 May;10(3):217-24. doi: 10.2353/jmoldx.2008.070100. Epub 2008 Apr 10. J Mol Diagn. 2008. PMID: 18403612 Free PMC article.
-
DNA pyrosequencing-based bacterial pathogen identification in a pediatric hospital setting.J Clin Microbiol. 2007 Sep;45(9):2985-92. doi: 10.1128/JCM.00630-07. Epub 2007 Jul 25. J Clin Microbiol. 2007. PMID: 17652476 Free PMC article.
-
Focused, high accuracy 5-methylcytosine quantitation with base resolution by benchtop next-generation sequencing.Epigenetics Chromatin. 2013 Oct 11;6(1):33. doi: 10.1186/1756-8935-6-33. Epigenetics Chromatin. 2013. PMID: 24279302 Free PMC article.
-
A multi-site study using high-resolution HLA genotyping by next generation sequencing.Tissue Antigens. 2011 Mar;77(3):206-17. doi: 10.1111/j.1399-0039.2010.01606.x. Tissue Antigens. 2011. PMID: 21299525 Free PMC article.
-
What Next? The Next Transit from Biology to Diagnostics: Next Generation Sequencing for Immunogenetics.Transfus Med Hemother. 2011 Oct;38(5):308-317. doi: 10.1159/000332433. Epub 2011 Sep 25. Transfus Med Hemother. 2011. PMID: 22670120 Free PMC article.
References
-
- Ronaghi M, Karamohamed S, Pettersson B, Uhlen M, Nyren P. Real-time DNA sequencing using detection of pyrophosphate release. Analytical Biochemistry. 1996;242:84–89. - PubMed
-
- Ronaghi M, Uhlen M, Nyren P. A sequencing method based on real-time pyrophosphate. Science. 1998;281:363. - PubMed
-
- Ahmadian A, Gharizadeh B, Gustafsson AC, Sterky F, Nyren P, Uhlen M, Lundeberg J. Single-nucleotide polymorphism analysis by Pyrosequencing. Anal Biochem. 2000;280:103–110. - PubMed
-
- Fakhrai-Rad H, Pourmand N, Ronaghi M. Pyrosequencing: an accurate detection platform for single nucleotide polymorphisms. Hum Mutat. 2002;19:479–485. - PubMed
-
- Ronaghi M. Pyrosequencing for SNP genotyping. Methods Mol Biol. 2003;212:189–195. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

