APOBEC3G multimers are recruited to the plasma membrane for packaging into human immunodeficiency virus type 1 virus-like particles in an RNA-dependent process requiring the NC basic linker
- PMID: 17344295
- PMCID: PMC1900209
- DOI: 10.1128/JVI.02237-06
APOBEC3G multimers are recruited to the plasma membrane for packaging into human immunodeficiency virus type 1 virus-like particles in an RNA-dependent process requiring the NC basic linker
Abstract
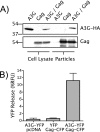
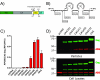
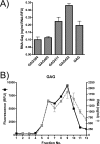
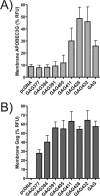
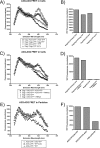
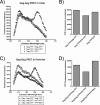
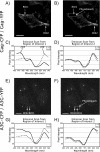
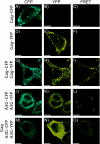
APOBEC3G is an endogenous host restriction factor that inhibits human immunodeficiency virus (HIV) replication. The antiviral activity of APOBEC3G is dependent upon its incorporation into the virus particle. The mechanisms governing incorporation of APOBEC3G into virus particles are not completely understood. In particular, some investigators have reported that APOBEC3G interacts directly with the nucleocapsid (NC) subunit of Gag, while others have found that an RNA intermediate is required for Gag-APOBEC3G interactions. In this study, we confirmed the RNA dependence of APOBEC3G packaging and performed detailed mapping of the determinants within NC that are required for virion incorporation. Surprisingly, APOBEC3G packaging did not correlate well with the presence of the N-terminal "I," or interaction, domain within NC. Specifically, Gag constructs containing only the N-terminal region of NC packaged minimal amounts of APOBEC3G, while significant levels of APOBEC3G packaging were achieved with Gag constructs containing the basic linker region of NC. Furthermore, membrane-binding experiments revealed that the basic linker region was essential for the membrane association of APOBEC3G in a Gag-APOBEC3G complex. Fluorescence resonance energy transfer was detected between labeled APOBEC3G in cells and in particles, indicating that APOBEC3G is packaged as a multimer that is bound to packaged RNA. Regions of APOBEC3G-Gag colocalization at the plasma membrane were detected that were distinct from the punctate cytoplasmic bodies where APOBEC3G accumulates within the cell. Together, our results indicate that APOBEC3G multimerizes in an RNA-dependent fashion and that RNA-APOBEC3G multimers are recruited to the plasma membrane and subsequently into virion particles by Gag.
Figures


References
-
- Alce, T. M., and W. Popik. 2004. APOBEC3G is incorporated into virus-like particles by a direct interaction with HIV-1 Gag nucleocapsid protein. J. Biol. Chem. 279:34083-34086. - PubMed
-
- Cen, S., F. Guo, M. Niu, J. Saadatmand, J. Deflassieux, and L. Kleiman. 2004. The interaction between HIV-1 Gag and APOBEC3G. J. Biol. Chem. 279:33177-33184. - PubMed
-
- Chiu, Y. L., V. B. Soros, J. F. Kreisberg, K. Stopak, W. Yonemoto, and W. C. Greene. 2005. Cellular APOBEC3G restricts HIV-1 infection in resting CD4+ T cells. Nature 435:108-114. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

