GGA2- and ubiquitin-dependent trafficking of Arn1, the ferrichrome transporter of Saccharomyces cerevisiae
- PMID: 17344478
- PMCID: PMC1855028
- DOI: 10.1091/mbc.e06-09-0861
GGA2- and ubiquitin-dependent trafficking of Arn1, the ferrichrome transporter of Saccharomyces cerevisiae
Abstract
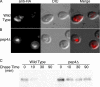
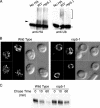
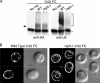
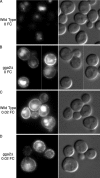
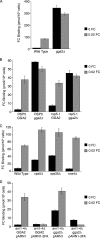
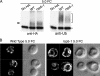
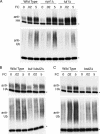
The intracellular trafficking of Arn1, a ferrichrome transporter in Saccharomyces cerevisiae, is controlled in part by the binding of ferrichrome to the transporter. In the absence of ferrichrome, Arn1 is sorted directly from the Golgi to endosomes. Ferrichrome binding triggers the redistribution of Arn1 to the plasma membrane, whereas ferrichrome transport is associated with the cycling of Arn1 between the plasma membrane and endosomes. Here, we report that the clathrin adaptor Gga2 and ubiquitination by the Rsp5 ubiquitin ligase are required for trafficking of Arn1. Gga2 was required for Golgi-to-endosomal trafficking of Arn1, which was sorted from endosomes to the vacuole for degradation. Trafficking into the vacuolar lumen was dependent on ubiquitination by Rsp5, but ubiquitination was not required for plasma membrane accumulation of Arn1 in the presence of ferrichrome. Retrograde trafficking via the retromer complex or Snx4 was also not required for plasma membrane accumulation. High concentrations of ferrichrome led to higher levels of ubiquitination of Arn1, but they did not induce degradation. Without this ubiquitination, Arn1 remained on the plasma membrane, where it was active for transport. Arn1 was preferentially modified with polyubiquitin chains on a cluster of lysine residues at the amino terminus of the transporter.
Figures
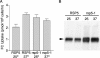

Similar articles
-
Gga2 mediates sequential ubiquitin-independent and ubiquitin-dependent steps in the trafficking of ARN1 from the trans-Golgi network to the vacuole.J Biol Chem. 2009 Aug 28;284(35):23830-41. doi: 10.1074/jbc.M109.030015. Epub 2009 Jul 1. J Biol Chem. 2009. PMID: 19574226 Free PMC article.
-
Phosphatidylserine is involved in the ferrichrome-induced plasma membrane trafficking of Arn1 in Saccharomyces cerevisiae.J Biol Chem. 2010 Dec 10;285(50):39564-73. doi: 10.1074/jbc.M110.177055. Epub 2010 Oct 5. J Biol Chem. 2010. PMID: 20923770 Free PMC article.
-
A receptor domain controls the intracellular sorting of the ferrichrome transporter, ARN1.EMBO J. 2005 Mar 9;24(5):952-62. doi: 10.1038/sj.emboj.7600579. Epub 2005 Feb 17. EMBO J. 2005. PMID: 15719020 Free PMC article.
-
The ubiquitin code of yeast permease trafficking.Trends Cell Biol. 2010 Apr;20(4):196-204. doi: 10.1016/j.tcb.2010.01.004. Trends Cell Biol. 2010. PMID: 20138522 Review.
-
Versatile role of the yeast ubiquitin ligase Rsp5p in intracellular trafficking.Biochem Soc Trans. 2008 Oct;36(Pt 5):791-6. doi: 10.1042/BST0360791. Biochem Soc Trans. 2008. PMID: 18793138 Review.
Cited by
-
Direct binding of the Kex2p cytosolic tail to the VHS domain of yeast Gga2p facilitates TGN to prevacuolar compartment transport and is regulated by phosphorylation.Mol Biol Cell. 2013 Feb;24(4):495-509. doi: 10.1091/mbc.E12-11-0843. Mol Biol Cell. 2013. PMID: 23408788 Free PMC article.
-
Regulation of cation balance in Saccharomyces cerevisiae.Genetics. 2013 Mar;193(3):677-713. doi: 10.1534/genetics.112.147207. Genetics. 2013. PMID: 23463800 Free PMC article. Review.
-
Gga2 mediates sequential ubiquitin-independent and ubiquitin-dependent steps in the trafficking of ARN1 from the trans-Golgi network to the vacuole.J Biol Chem. 2009 Aug 28;284(35):23830-41. doi: 10.1074/jbc.M109.030015. Epub 2009 Jul 1. J Biol Chem. 2009. PMID: 19574226 Free PMC article.
-
Glucose-induced ubiquitylation and endocytosis of the yeast Jen1 transporter: role of lysine 63-linked ubiquitin chains.J Biol Chem. 2009 Jul 17;284(29):19228-36. doi: 10.1074/jbc.M109.008318. Epub 2009 May 11. J Biol Chem. 2009. PMID: 19433580 Free PMC article.
-
Dual sorting of the Saccharomyces cerevisiae vacuolar protein Sna4p.Eukaryot Cell. 2009 Mar;8(3):278-86. doi: 10.1128/EC.00363-08. Epub 2009 Jan 23. Eukaryot Cell. 2009. PMID: 19168755 Free PMC article.
References
-
- Barriere H., Nemes C., Lechardeur D., Khan-Mohammad M., Fruh K., Lukacs G. L. Molecular basis of oligoubiquitin-dependent internalization of membrane proteins in mammalian cells. Traffic. 2006;7:282–297. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

