Bone marrow mesenchymal stem cells are abnormal in multiple myeloma
- PMID: 17344918
- PMCID: PMC2346535
- DOI: 10.1038/sj.leu.2404621
Bone marrow mesenchymal stem cells are abnormal in multiple myeloma
Abstract
Recent literature suggested that cells of the microenvironment of tumors could be abnormal as well. To address this hypothesis in multiple myeloma (MM), we studied bone marrow mesenchymal stem cells (BMMSCs), the only long-lived cells of the bone marrow microenvironment, by gene expression profiling and phenotypic and functional studies in three groups of individuals: patients with MM, patients with monoclonal gamopathy of undefined significance (MGUS) and healthy age-matched subjects. Gene expression profile independently classified the BMMSCs of these individuals in a normal and in an MM group. MGUS BMMSCs were interspersed between these two groups. Among the 145 distinct genes differentially expressed in MM and normal BMMSCs, 46% may account for a tumor-microenvironment cross-talk. Known soluble factors implicated in MM pathophysiologic features (i.e. IL (interleukin)-6, DKK1) were revealed and new ones were found which are involved in angiogenesis, osteogenic differentiation or tumor growth. In particular, GDF15 was found to induce dose-dependent growth of MOLP-6, a stromal cell-dependent myeloma cell line. Functionally, MM BMMSCs induced an overgrowth of MOLP-6, and their capacity to differentiate into an osteoblastic lineage was impaired. Thus, MM BMMSCs are abnormal and could create a very efficient niche to support the survival and proliferation of the myeloma cells.
Figures
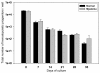

 ). Data were obtained from U133 2plus microarrays (Affymetrix) as described in Materials and Methods. Analysis was performed on 28746 probe sets declared as present by the call detection algorithm. The variation coefficient (VC), defined as the ratio of the SD to the mean of signal values for the considered probe set, was used to select genes incorporated in the PCA. By varying VC from 0% to 100%, the normal and MM BMMSC clusters can be more or less separated. At a VC of 60% the separation was the best and includes ~ 2,000 genes. The boxed scheme corresponds to the cluster analysis performed on the same set of data with Cluster and TreeView softwares (
). Data were obtained from U133 2plus microarrays (Affymetrix) as described in Materials and Methods. Analysis was performed on 28746 probe sets declared as present by the call detection algorithm. The variation coefficient (VC), defined as the ratio of the SD to the mean of signal values for the considered probe set, was used to select genes incorporated in the PCA. By varying VC from 0% to 100%, the normal and MM BMMSC clusters can be more or less separated. At a VC of 60% the separation was the best and includes ~ 2,000 genes. The boxed scheme corresponds to the cluster analysis performed on the same set of data with Cluster and TreeView softwares ( ) were added to the analysis.
) were added to the analysis.
 ). Data were obtained from U133 2plus microarrays (Affymetrix) as described in Materials and Methods. Analysis was performed on 28746 probe sets declared as present by the call detection algorithm. The variation coefficient (VC), defined as the ratio of the SD to the mean of signal values for the considered probe set, was used to select genes incorporated in the PCA. By varying VC from 0% to 100%, the normal and MM BMMSC clusters can be more or less separated. At a VC of 60% the separation was the best and includes ~ 2,000 genes. The boxed scheme corresponds to the cluster analysis performed on the same set of data with Cluster and TreeView softwares (
). Data were obtained from U133 2plus microarrays (Affymetrix) as described in Materials and Methods. Analysis was performed on 28746 probe sets declared as present by the call detection algorithm. The variation coefficient (VC), defined as the ratio of the SD to the mean of signal values for the considered probe set, was used to select genes incorporated in the PCA. By varying VC from 0% to 100%, the normal and MM BMMSC clusters can be more or less separated. At a VC of 60% the separation was the best and includes ~ 2,000 genes. The boxed scheme corresponds to the cluster analysis performed on the same set of data with Cluster and TreeView softwares ( ) were added to the analysis.
) were added to the analysis.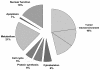
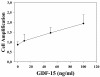
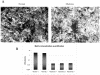
References
-
- Liotta LA, Kohn EC. The microenvironment of the tumour-host interface. Nature. 2001;411:375–379. - PubMed
-
- Maffini MV, Soto AM, Calabro JM, Ucci AA, Sonnenschein C. The stroma as a crucial target in rat mammary gland carcinogenesis. J Cell Sci. 2004;117:1495–1502. - PubMed
-
- Allinen M, Beroukhim R, Cai L, et al. Molecular characterization of the tumor microenvironment in breast cancer. Cancer Cell. 2004;6:17–32. - PubMed
-
- Attal M, Harousseau JL, Stoppa AM, et al. A prospective, randomized trial of autologous bone marrow transplantation and chemotherapy in multiple myeloma. Intergroupe Francais du Myelome. N Engl J Med. 1996;335:91–97. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

