Multilocus perspectives on the monophyly and phylogeny of the order Charadriiformes (Aves)
- PMID: 17346347
- PMCID: PMC1838420
- DOI: 10.1186/1471-2148-7-35
Multilocus perspectives on the monophyly and phylogeny of the order Charadriiformes (Aves)
Abstract
Background: The phylogeny of shorebirds (Aves: Charadriiformes) and their putative sister groups was reconstructed using approximately 5 kilobases of data from three nuclear loci and two mitochondrial genes, and compared to that based on two other nuclear loci.
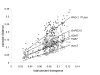
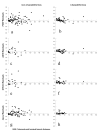
Results: Charadriiformes represent a monophyletic group that consists of three monophyletic suborders Lari (i.e., Laridae [including Sternidae and Rynchopidae], Stercorariidae, Alcidae, Glareolidae, Dromadidae, and Turnicidae), Scolopaci (i.e., Scolopacidae [including Phalaropidae], Jacanidae, Rostratulidae, Thinocoridae, Pedionomidae), and Charadrii (i.e., Burhinidae, Chionididae, Charadriidae, Haematopodidae, Recurvirostridae, and presumably Ibidorhynchidae). The position of purported "gruiform" buttonquails within Charadriiformes is confirmed. Skimmers are most likely sister to terns alone, and plovers may be paraphyletic with respect to oystercatchers and stilts. The Egyptian Plover is not a member of the Glareolidae, but is instead relatively basal among Charadrii. None of the putative sisters of Charadriiformes were recovered as such.
Conclusion: Hypotheses of non-monophyly and sister relationships of shorebirds are tested by multilocus analysis. The monophyly of and interfamilial relationships among shorebirds are confirmed and refined. Lineage-specific differences in evolutionary rates are more consistent across loci in shorebirds than other birds and may contribute to the congruence of locus-specific phylogenetic estimates in shorebirds.
Figures
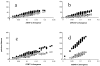

References
-
- Van Tuinen M, Waterhouse D, Dyke GJ. Avian molecular systematics on the rebound: a fresh look at modern shorebird phylogenetic relationships. J Avian Biol. 2004;35:191–194. doi: 10.1111/j.0908-8857.2004.03362.x. - DOI
-
- Strauch JG., Jr. The phylogeny of Charadriiformes (Aves): a new estimate using the method of character compatibility analysis. Trans Zool Soc London. 1978;34:263–345.
-
- Chu PC. Phylogenetic reanalysis of Strauch’s osteological data set for the Charadriiformes. Condor. 1995;97:174–196. doi: 10.2307/1368995. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

