Multiple origins of replication contribute to a discontinuous pattern of DNA synthesis across the T4 genome during infection
- PMID: 17346743
- PMCID: PMC1934900
- DOI: 10.1016/j.jmb.2007.02.008
Multiple origins of replication contribute to a discontinuous pattern of DNA synthesis across the T4 genome during infection
Abstract
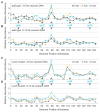
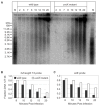
Chromosomes provide a template for a number of DNA transactions, including replication and transcription, but the dynamic interplay between these activities is poorly understood at the genomic level. The bacteriophage T4 has long served as a model for the study of DNA replication, transcription, and recombination, and should be an excellent model organism in which to integrate in vitro biochemistry into a chromosomal context. As a first step in characterizing the dynamics of chromosomal transactions during T4 infection, we have employed a unique set of macro array strategies to identify the origins of viral DNA synthesis and monitor the actual accumulation of nascent DNA across the genome in real time. We show that T4 DNA synthesis originates from at least five discrete loci within a single population of infected cells, near oriA, oriC, oriE, oriF, and oriG, the first direct evidence of multiple, active origins within a single population of infected cells. Although early T4 DNA replication is initiated at defined origins, continued synthesis requires viral recombination. The relationship between these two modes of replication during infection has not been well understood, but we observe that the switch between origin and recombination-mediated replication is dependent on the number of infecting viruses. Finally, we demonstrate that the nascent DNAs produced from origin loci are regulated spatially and temporally, leading to the accumulation of multiple, short DNAs near the origins, which are presumably used to prime subsequent recombination-mediated replication. These results provide the foundation for the future characterization of the molecular dynamics that contribute to T4 genome function and evolution and may provide insights into the replication of other multi origin chromosomes.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

