Biosynthesis of ether-type polar lipids in archaea and evolutionary considerations
- PMID: 17347520
- PMCID: PMC1847378
- DOI: 10.1128/MMBR.00033-06
Biosynthesis of ether-type polar lipids in archaea and evolutionary considerations
Abstract
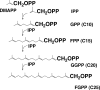
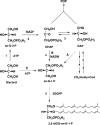
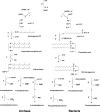
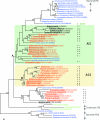
This review deals with the in vitro biosynthesis of the characteristics of polar lipids in archaea along with preceding in vivo studies. Isoprenoid chains are synthesized through the classical mevalonate pathway, as in eucarya, with minor modifications in some archaeal species. Most enzymes involved in the pathway have been identified enzymatically and/or genomically. Three of the relevant enzymes are found in enzyme families different from the known enzymes. The order of reactions in the phospholipid synthesis pathway (glycerophosphate backbone formation, linking of glycerophosphate with two radyl chains, activation by CDP, and attachment of common polar head groups) is analogous to that of bacteria. sn-Glycerol-1-phosphate dehydrogenase is responsible for the formation of the sn-glycerol-1-phosphate backbone of phospholipids in all archaea. After the formation of two ether bonds, CDP-archaeol acts as a common precursor of various archaeal phospholipid syntheses. Various phospholipid-synthesizing enzymes from archaea and bacteria belong to the same large CDP-alcohol phosphatidyltransferase family. In short, the first halves of the phospholipid synthesis pathways play a role in synthesis of the characteristic structures of archaeal and bacterial phospholipids, respectively. In the second halves of the pathways, the polar head group-attaching reactions and enzymes are homologous in both domains. These are regarded as revealing the hybrid nature of phospholipid biosynthesis. Precells proposed by Wächtershäuser are differentiated into archaea and bacteria by spontaneous segregation of enantiomeric phospholipid membranes (with sn-glycerol-1-phosphate and sn-glycerol-3-phosphate backbones) and the fusion and fission of precells. Considering the nature of the phospholipid synthesis pathways, we here propose that common phospholipid polar head groups were present in precells before the differentiation into archaea and bacteria.
Figures







References
-
- Bachhawat, N., and S. C. Mande. 2000. Complex evolution of the inositol-1-phosphate synthase gene among archaea and eubacteria. Trends Genet. 16:111-113. - PubMed
-
- Baxter, R. M., and N. E. Gibbons. 1954. The glycerol dehydrogenases of Pseudomonas salinaria, Vibrio costicolus, and Escherichia coli in relation to bacterial halophilism. Can. J. Biochem. Physiol. 32:206-217. - PubMed
-
- Beisenherz, G. 1955. Triose phosphate isomerase from calf muscle. Methods Enzymol. 1:387-391.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

