Colicin biology
- PMID: 17347522
- PMCID: PMC1847374
- DOI: 10.1128/MMBR.00036-06
Colicin biology
Abstract
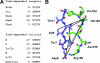
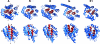
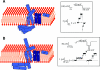
Colicins are proteins produced by and toxic for some strains of Escherichia coli. They are produced by strains of E. coli carrying a colicinogenic plasmid that bears the genetic determinants for colicin synthesis, immunity, and release. Insights gained into each fundamental aspect of their biology are presented: their synthesis, which is under SOS regulation; their release into the extracellular medium, which involves the colicin lysis protein; and their uptake mechanisms and modes of action. Colicins are organized into three domains, each one involved in a different step of the process of killing sensitive bacteria. The structures of some colicins are known at the atomic level and are discussed. Colicins exert their lethal action by first binding to specific receptors, which are outer membrane proteins used for the entry of specific nutrients. They are then translocated through the outer membrane and transit through the periplasm by either the Tol or the TonB system. The components of each system are known, and their implication in the functioning of the system is described. Colicins then reach their lethal target and act either by forming a voltage-dependent channel into the inner membrane or by using their endonuclease activity on DNA, rRNA, or tRNA. The mechanisms of inhibition by specific and cognate immunity proteins are presented. Finally, the use of colicins as laboratory or biotechnological tools and their mode of evolution are discussed.
Figures




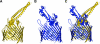










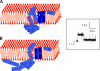

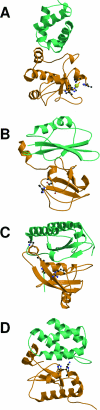



References
-
- Abergel, C., E. Bouveret, J. M. Claverie, K. Brown, A. Rigal, C. Lazdunski, and H. Bénédetti. 1999. Structure of the Escherichia coli TolB protein determined by MAD methods at 1.95 A resolution. Structure 7:1291-1300. - PubMed
-
- Amati, P. 1964. Vegetative multiplication of colicinogenic factors after induction in Escherichia coli. J. Mol. Biol. 78:239-246. - PubMed
-
- Anderluh, G., I. Gokce, and J. H. Lakey. 2004. A natively unfolded toxin domain uses its receptor as a folding template. J. Biol. Chem. 279:22002-22009. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

