Rad50 adenylate kinase activity regulates DNA tethering by Mre11/Rad50 complexes
- PMID: 17349953
- PMCID: PMC3050042
- DOI: 10.1016/j.molcel.2007.01.028
Rad50 adenylate kinase activity regulates DNA tethering by Mre11/Rad50 complexes
Abstract
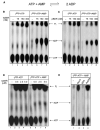
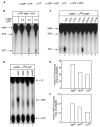
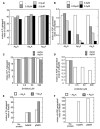
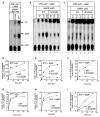
Mre11 and Rad50 are the catalytic components of a highly conserved DNA repair complex that functions in many aspects of DNA metabolism involving double-strand breaks. The ATPase domains in Rad50 are related to the ABC transporter family of ATPases, previously shown to share structural similarities with adenylate kinases. Here we demonstrate that Mre11/Rad50 complexes from three organisms catalyze the reversible adenylate kinase reaction in vitro. Mutation of the conserved signature motif reduces the adenylate kinase activity of Rad50 but does not reduce ATP hydrolysis. This mutant resembles a rad50 null strain with respect to meiosis and telomere maintenance in S. cerevisiae, correlating adenylate kinase activity with in vivo functions. An adenylate kinase inhibitor blocks Mre11/Rad50-dependent DNA tethering in vitro and in cell-free extracts, indicating that adenylate kinase activity by Mre11/Rad50 promotes DNA-DNA associations. We propose a model for Rad50 that incorporates both ATPase and adenylate kinase reactions as critical activities that regulate Rad50 functions.
Figures
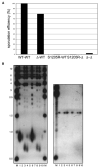
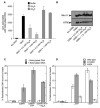
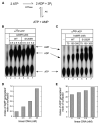
Comment in
-
Learning our ABCs: Rad50 directs MRN repair functions via adenylate kinase activity from the conserved ATP binding cassette.Mol Cell. 2007 Mar 23;25(6):789-91. doi: 10.1016/j.molcel.2007.03.004. Mol Cell. 2007. PMID: 17386254
References
-
- Adams A, Gottschling DE, Kaiser CA, Stearns T. Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1997.
-
- Alani E, Padmore R, Kleckner N. Analysis of wild-type and rad50 mutants of yeast suggests an intimate relationship between meiotic chromosome synapsis and recombination. Cell. 1990;61:419–436. - PubMed
-
- Chen L, Trujillo K, Ramos W, Sung P, Tomkinson AE. Promotion of Dnl4-catalyzed DNA end-joining by the Rad50/Mre11/Xrs2 and Hdf1/Hdf2 complexes. Mol Cell. 2001;8:1105–1115. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

