Luminex detection of fecal indicators in river samples, marine recreational water, and beach sand
- PMID: 17350051
- PMCID: PMC1950674
- DOI: 10.1016/j.marpolbul.2006.12.018
Luminex detection of fecal indicators in river samples, marine recreational water, and beach sand
Abstract
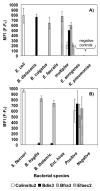
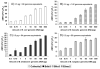
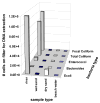
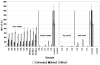
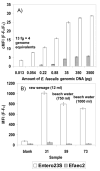
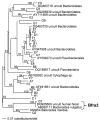
Research to understand and remediate coastal pollution is moving toward a multitiered approach in which traditional enumeration of fecal indicators is accompanied by molecular analysis of a variety of targets. Technology that rapidly detects multiple microbial contaminants would benefit from such an approach. The Luminex 100 system is a suspension array that assays multiple analytes rapidly in a single well of a microtiter plate. The ability of the system to simultaneously detect multiple fecal indicating bacteria in environmental samples was tested. Primer/probe sets were designed to simultaneously detect the following fecal indicators: the Bacteroides fragilis group, Enterococcus spp., Escherichia coli and Shigella spp., Bacteroides distasonis, and Ent. faecalis. Specificity and sensitivity of the Luminex probes was tested against laboratory cultures. In addition, sequencing, culture plate testing, and specificity testing with environmental isolates were steps taken to validate the function of the assay with environmental samples. Luminex response to cultures and to environmental samples was consistent with sequencing results, suggesting that the technology has the potential to simultaneously detect multiple targets for coastal water quality applications, particularly as progress is made to efficiently extract DNA from water and sediment matrices.
Figures

References
-
- Allsop K, Stickler DJ. An assessment of Bacteroides fragilis group organisms as indicators of human faecal pollution. Journal of Applied Bacteriology. 1985;58:95–99. - PubMed
-
- Alm EW, Burke J, Hagan E. Persistence and potential growth of the fecal indicator bacteria, Escherichia coli in shoreline sand at Lake Huron. J Great Lakes Res. 2006;32:401–405.
-
- Bernhard AE, Colbert D, McManus J, Field KG. Microbial community dynamics based on 16S rRNA gene profiles in a Pacific Northwest estuary and its tributaries. FEMS Microbiology Ecology. 2005;52:115–128. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

