An Nkx2-5/Bmp2/Smad1 negative feedback loop controls heart progenitor specification and proliferation
- PMID: 17350578
- PMCID: PMC2092439
- DOI: 10.1016/j.cell.2007.01.042
An Nkx2-5/Bmp2/Smad1 negative feedback loop controls heart progenitor specification and proliferation
Abstract
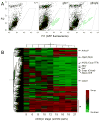
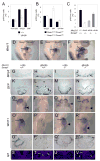
During heart development the second heart field (SHF) provides progenitor cells for most cardiomyocytes and expresses the homeodomain factor Nkx2-5. We now show that feedback repression of Bmp2/Smad1 signaling by Nkx2-5 critically regulates SHF proliferation and outflow tract (OFT) morphology. In the cardiac fields of Nkx2-5 mutants, genes controlling cardiac specification (including Bmp2) and maintenance of the progenitor state were upregulated, leading initially to progenitor overspecification, but subsequently to failed SHF proliferation and OFT truncation. In Smad1 mutants, SHF proliferation and deployment to the OFT were increased, while Smad1 deletion in Nkx2-5 mutants rescued SHF proliferation and OFT development. In Nkx2-5 hypomorphic mice, which recapitulate human congenital heart disease (CHD), OFT anomalies were also rescued by Smad1 deletion. Our findings demonstrate that Nkx2-5 orchestrates the transition between periods of cardiac induction, progenitor proliferation, and OFT morphogenesis via a Smad1-dependent negative feedback loop, which may be a frequent molecular target in CHD.
Figures





References
-
- Biben C, Harvey RP. Homeodomain factor Nkx2-5 controls left-right asymmetric expression of bHLH eHand during murine heart development. Genes Dev. 1997;11:1357–1369. - PubMed
-
- Biben C, Weber R, Kesteven S, Stanley E, McDonald L, Elliott DA, Barnett L, Koentgen F, Robb L, Feneley M, Harvey RP. Cardiac septal and valvular dysmorphogenesis in mice heterozygous for mutations in the homeobox gene Nkx2-5. Circ Res. 2000;87:888–895. - PubMed
-
- Buckingham M, Meilhac S, Zaffran S. Building the mammalian heart from two sources of myocardial cells. Nat Rev Genet. 2005;6:826–835. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

