The genome of Hyperthermus butylicus: a sulfur-reducing, peptide fermenting, neutrophilic Crenarchaeote growing up to 108 degrees C
- PMID: 17350933
- PMCID: PMC2686385
- DOI: 10.1155/2007/745987
The genome of Hyperthermus butylicus: a sulfur-reducing, peptide fermenting, neutrophilic Crenarchaeote growing up to 108 degrees C
Abstract
Hyperthermus butylicus, a hyperthermophilic neutrophile and anaerobe, is a member of the archaeal kingdom Crenarchaeota. Its genome consists of a single circular chromosome of 1,667,163 bp with a 53.7% G+C content. A total of 1672 genes were annotated, of which 1602 are protein-coding, and up to a third are specific to H. butylicus. In contrast to some other crenarchaeal genomes, a high level of GUG and UUG start codons are predicted. Two cdc6 genes are present, but neither could be linked unambiguously to an origin of replication. Many of the predicted metabolic gene products are associated with the fermentation of peptide mixtures including several peptidases with diverse specificities, and there are many encoded transporters. Most of the sulfur-reducing enzymes, hydrogenases and electron-transfer proteins were identified which are associated with energy production by reducing sulfur to H(2)S. Two large clusters of regularly interspaced repeats (CRISPRs) are present, one of which is associated with a crenarchaeal-type cas gene superoperon; none of the spacer sequences yielded good sequence matches with known archaeal chromosomal elements. The genome carries no detectable transposable or integrated elements, no inteins, and introns are exclusive to tRNA genes. This suggests that the genome structure is quite stable, possibly reflecting a constant, and relatively uncompetitive, natural environment.
Figures
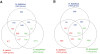
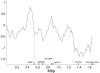

References
-
- Bendtsen J.D., Nielsen H., Von heijne G., Brunak S. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 2004;340:783–795. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

