Unfolding of beta-sheet proteins in SDS
- PMID: 17351005
- PMCID: PMC1853130
- DOI: 10.1529/biophysj.106.101238
Unfolding of beta-sheet proteins in SDS
Abstract
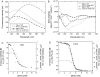
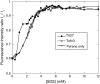
Beta-sheet proteins are particularly resistant to denaturation by sodium dodecyl sulfate (SDS). Here we compare unfolding of two beta-sandwich proteins TNfn3 and TII27 in SDS. The two proteins show different surface electrostatic potential. Correspondingly, TII27 unfolds below the critical micelle concentration via the formation of hemimicelles on the protein surface, whereas TNfn3 only unfolds around the critical micelle concentration. Isothermal titration calorimetry confirms that unfolding of TII27 sets in at lower SDS concentrations, although the total number of bound SDS molecules is similar at the end of unfolding. In mixed micelles with the nonionic detergent dodecyl maltoside, where the concentration of monomeric SDS is insignificant, the behavior of the two proteins converges. TII27 unfolds more slowly than TNfn3 in SDS and follows a two-mode behavior. Additionally TNfn3 shows inhibition of SDS unfolding at intermediate SDS concentrations. Mutagenic analysis suggests that the overall unfolding mechanism is similar to that observed in denaturant for both proteins. Our data confirm the kinetic robustness of beta-sheet proteins toward SDS. We suggest this is related to the inability of SDS to induce significant amounts of alpha-helix structure in these proteins as part of the denaturation process, forcing the protein to denature by global rather than local unfolding.
Figures






References
-
- La Mesa, C. 2005. Polymer-surfactant and protein-surfactant interactions. J. Coll. Int. Sci. 286:148–157. - PubMed
-
- Timasheff, S. 2002. Protein hydration, thermodynamic binding, and preferential hydration. Biochemistry. 41:13473–13482. - PubMed
-
- Kirk, O., T. V. Borchert, and C. C. Fuglsang. 2002. Industrial enzyme applications. Curr. Opin. Biotechnol. 13:345–351. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

