A WUSCHEL-LIKE HOMEOBOX gene represses a YABBY gene expression required for rice leaf development
- PMID: 17351053
- PMCID: PMC1913789
- DOI: 10.1104/pp.107.095737
A WUSCHEL-LIKE HOMEOBOX gene represses a YABBY gene expression required for rice leaf development
Abstract
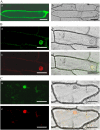
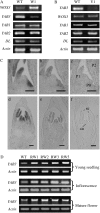
YABBY and WUSCHEL-LIKE HOMEOBOX (WOX) genes have been shown to play important roles in lateral organ formation and meristem function. Here, we report the characterization of functional relationship between rice (Oryza sativa) YAB3 and WOX3 in rice leaf development. Rice YAB3 is closely related to maize (Zea mays) ZmYAB14 and Arabidopsis (Arabidopsis thaliana) FILAMENTOUS FLOWER (FIL), whereas rice WOX3 is highly conserved with maize narrow sheath1 (NS1) and NS2 and Arabidopsis PRESSED FLOWER (PRS). In situ hybridization experiments revealed that the expression of both genes was excluded from the shoot apical meristem, but the transcripts were detected in leaf primordia, young leaves, and reproductive organs without any polar distribution. The function of the two genes was studied by both overexpression and RNA interference (RNAi) in transgenic rice. YAB3 RNAi induced twisted and knotted leaves lacking specialized structures such as ligule and auricles, while no phenotypic change was observed in YAB3 overexpression plants, suggesting that rice YAB3 may be required for leaf cell growth and differentiation. Overexpression of WOX3 repressed YAB3 and showed a YAB3 RNAi phenotype. The expression of class I KNOTTED-LIKE HOMEOBOX (KNOX) genes was ectopically induced in leaves of YAB3 RNAi or WOX3 overexpression plants. Data from inducible WOX3 expression and DNA-protein interaction assays suggested that WOX3 acted as a transcriptional repressor of YAB3. These data reveal a regulatory network involving YAB3, WOX3, and KNOX genes required for rice leaf development.
Figures






References
-
- Bowman JL (2000) The YABBY gene family and abaxial cell fate. Curr Opin Plant Biol 3 17–22 - PubMed
-
- Bowman JL, Smyth DR (1999) CRABS CLAW, a gene that regulates carpel and nectary development in Arabidopsis, encodes a novel protein with zinc finger and helix-loop-helix domains. Development 126 2387–2396 - PubMed
-
- Byrne ME, Barley R, Curtis M, Arroyo JM, Dunham M, Hudson A, Martienssen RA (2000) Asymmetric leaves1 mediates leaf patterning and stem cell function in Arabidopsis. Nature 408 967–971 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

