Magnaporthe grisea infection triggers RNA variation and antisense transcript expression in rice
- PMID: 17351054
- PMCID: PMC1913787
- DOI: 10.1104/pp.107.095653
Magnaporthe grisea infection triggers RNA variation and antisense transcript expression in rice
Abstract
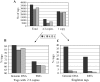
Rice blast disease, caused by the fungal pathogen Magnaporthe grisea, is an excellent model system to study plant-fungal interactions and host defense responses. In this study, comprehensive analysis of the rice (Oryza sativa) transcriptome after M. grisea infection was conducted using robust-long serial analysis of gene expression. A total of 83,382 distinct 21-bp robust-long serial analysis of gene expression tags were identified from 627,262 individual tags isolated from the resistant (R), susceptible (S), and control (C) libraries. Sequence analysis revealed that the tags in the R and S libraries had a significant reduced matching rate to the rice genomic and expressed sequences in comparison to the C library. The high level of one-nucleotide mismatches of the R and S library tags was due to nucleotide conversions. The A-to-G and U-to-C nucleotide conversions were the most predominant types, which were induced in the M. grisea-infected plants. Reverse transcription-polymerase chain reaction analysis showed that expression of the adenine deaminase and cytidine deaminase genes was highly induced after inoculation. In addition, many antisense transcripts were induced in infected plants and expression of four antisense transcripts was confirmed by strand-specific reverse transcription-polymerase chain reaction. These results demonstrate that there is a series of dynamic and complex transcript modifications and changes in the rice transcriptome at the M. grisea early infection stages.
Figures




References
-
- Araya A, Zabaleta E, Blanc V, Begu D, Hernould M, Mouras A, Litvak S (1998) RNA editing in plant mitochondria, cytoplasmic male sterility and plant breeding. Biotechnol J 1 31–39
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

