A look inside HIV resistance through retroviral protease interaction maps
- PMID: 17352531
- PMCID: PMC1817660
- DOI: 10.1371/journal.pcbi.0030048
A look inside HIV resistance through retroviral protease interaction maps
Abstract
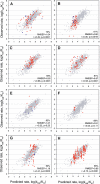
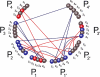
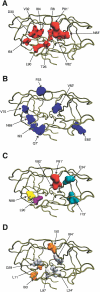
Retroviruses affect a large number of species, from fish and birds to mammals and humans, with global socioeconomic negative impacts. Here the authors report and experimentally validate a novel approach for the analysis of the molecular networks that are involved in the recognition of substrates by retroviral proteases. Using multivariate analysis of the sequence-based physiochemical descriptions of 61 retroviral proteases comprising wild-type proteases, natural mutants, and drug-resistant forms of proteases from nine different viral species in relation to their ability to cleave 299 substrates, the authors mapped the physicochemical properties and cross-dependencies of the amino acids of the proteases and their substrates, which revealed a complex molecular interaction network of substrate recognition and cleavage. The approach allowed a detailed analysis of the molecular-chemical mechanisms involved in substrate cleavage by retroviral proteases.
Conflict of interest statement
Figures

References
-
- Rosenberg N, Jolicoeur P. Retroviral pathogenesis. In: Coffin JM, Hughes SH, Varmus H, editors. Retroviruses. Cold Spring Harbor (New York): Cold Spring Harbor Laboratory Press; 1997. pp. 475–586. - PubMed
-
- Joint United Nations Programme on HIV/AIDS. 2006 Report on the Global AIDS Epidemic: Executive Summary. Vol. 13 Geneva (Switzerland): UNAIDS; 2006.
-
- Freire E. Designing drugs against heterogeneous targets. Nature Biotechnol. 2002;20:15–16. - PubMed
-
- Beck ZQ, Morris GM, Elder JH. Defining HIV-1 protease substrate selectivity. Curr Drug Targets Infect Disord. 2002;2:37–50. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources

