Differential regulation of Foxo3a target genes in erythropoiesis
- PMID: 17353275
- PMCID: PMC1900006
- DOI: 10.1128/MCB.01662-06
Differential regulation of Foxo3a target genes in erythropoiesis
Abstract
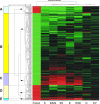
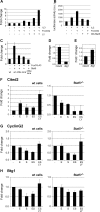
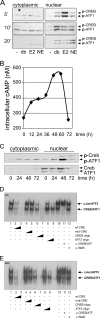
The cooperation of stem cell factor (SCF) and erythropoietin (Epo) is required to induce renewal divisions in erythroid progenitors, whereas differentiation to mature erythrocytes requires the presence of Epo only. Epo and SCF activate common signaling pathways such as the activation of protein kinase B (PKB) and the subsequent phosphorylation and inactivation of Foxo3a. In contrast, only Epo activates Stat5. Both Foxo3a and Stat5 promote erythroid differentiation. To understand the interplay of SCF and Epo in maintaining the balance between renewal and differentiation during erythroid development, we investigated differential Foxo3a target regulation by Epo and SCF. Expression profiling revealed that a subset of Foxo3a targets was not inhibited but was activated by Epo. One of these genes was Cited2. Transcriptional control of Epo/Foxo3a-induced Cited2 was studied and compared with that of the Epo-repressed Foxo3a target Btg1. We show that in response to Epo, the allegedly growth-inhibitory factor Foxo3a associates with the allegedly growth-stimulatory factor Stat5 in the nucleus, which is required for Epo-induced Cited2 expression. In contrast, Btg1 expression is controlled by the cooperation of Foxo3a with cyclic AMP- and Jun kinase-dependent Creb family members. Thus, Foxo3a not only is an effector of PKB but also integrates distinct signals to regulate gene expression in erythropoiesis.
Figures




Similar articles
-
Epo regulates erythroid proliferation and differentiation through distinct signaling pathways: implication for erythropoiesis and Friend virus-induced erythroleukemia.Oncogene. 2000 May 4;19(19):2296-304. doi: 10.1038/sj.onc.1203590. Oncogene. 2000. PMID: 10822380
-
Phosphorylation of forkhead transcription factors by erythropoietin and stem cell factor prevents acetylation and their interaction with coactivator p300 in erythroid progenitor cells.Oncogene. 2002 Feb 28;21(10):1556-62. doi: 10.1038/sj.onc.1205230. Oncogene. 2002. PMID: 11896584
-
FoxO3a regulates erythroid differentiation and induces BTG1, an activator of protein arginine methyl transferase 1.J Cell Biol. 2004 Jan 19;164(2):175-84. doi: 10.1083/jcb.200307056. J Cell Biol. 2004. PMID: 14734530 Free PMC article.
-
Control of erythropoiesis by erythropoietin and stem cell factor: a novel role for Bruton's tyrosine kinase.Cell Cycle. 2004 Jul;3(7):876-9. Epub 2004 Jul 2. Cell Cycle. 2004. PMID: 15254422 Review.
-
STAT5 as a Key Protein of Erythropoietin Signalization.Int J Mol Sci. 2021 Jul 1;22(13):7109. doi: 10.3390/ijms22137109. Int J Mol Sci. 2021. PMID: 34281163 Free PMC article. Review.
Cited by
-
Following Transcriptome to Uncover FOXO Biological Functions.Methods Mol Biol. 2019;1890:219-227. doi: 10.1007/978-1-4939-8900-3_18. Methods Mol Biol. 2019. PMID: 30414157 Free PMC article.
-
OutFOXOing disease and disability: the therapeutic potential of targeting FoxO proteins.Trends Mol Med. 2008 May;14(5):219-27. doi: 10.1016/j.molmed.2008.03.002. Epub 2008 Apr 9. Trends Mol Med. 2008. PMID: 18403263 Free PMC article. Review.
-
Clever cancer strategies with FoxO transcription factors.Cell Cycle. 2008 Dec 15;7(24):3829-39. doi: 10.4161/cc.7.24.7231. Epub 2008 Dec 21. Cell Cycle. 2008. PMID: 19066462 Free PMC article. Review.
-
Autophagy as a regulatory component of erythropoiesis.Int J Mol Sci. 2015 Feb 13;16(2):4083-94. doi: 10.3390/ijms16024083. Int J Mol Sci. 2015. PMID: 25689426 Free PMC article. Review.
-
FOXO3-mTOR metabolic cooperation in the regulation of erythroid cell maturation and homeostasis.Am J Hematol. 2014 Oct;89(10):954-63. doi: 10.1002/ajh.23786. Epub 2014 Jul 22. Am J Hematol. 2014. PMID: 24966026 Free PMC article.
References
-
- Abdel-Wahab, N., S. J. Wicks, R. M. Mason, and A. Chantry. 2002. Decorin suppresses transforming growth factor-beta-induced expression of plasminogen activator inhibitor-1 in human mesangial cells through a mechanism that involves Ca2+-dependent phosphorylation of Smad2 at serine-240. Biochem. J. 362:643-649. - PMC - PubMed
-
- Andrews, N. C., H. Erdjument-Bromage, M. B. Davidson, P. Tempst, and S. H. Orkin. 1993. Erythroid transcription factor NF-E2 is a haematopoietic-specific basic-leucine zipper protein. Nature 362:722-728. - PubMed
-
- Bakker, W. J., M. Blazquez-Domingo, A. Kolbus, J. Besooyen, P. Steinlein, H. Beug, P. J. Coffer, B. Lowenberg, M. Von Lindern, and T. B. Van Dijk. 2004. FoxO3a regulates erythroid differentiation and induces BTG1, an activator of protein arginine methyl transferase 1. J. Cell Biol. 164:175-184. - PMC - PubMed
-
- Bamforth, S. D., J. Braganca, C. R. Farthing, J. E. Schneider, C. Broadbent, A. C. Michell, K. Clarke, S. Neubauer, D. Norris, N. A. Brown, R. H. Anderson, and S. Bhattacharya. 2004. Cited2 controls left-right patterning and heart development through a Nodal-Pitx2c pathway. Nat. Genet. 36:1189-1196. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
