A U-turn motif-containing stem-loop in the coronavirus 5' untranslated region plays a functional role in replication
- PMID: 17353353
- PMCID: PMC1852815
- DOI: 10.1261/rna.261807
A U-turn motif-containing stem-loop in the coronavirus 5' untranslated region plays a functional role in replication
Abstract
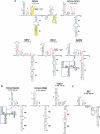
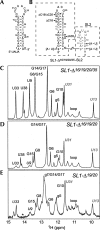
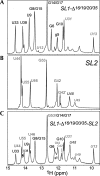
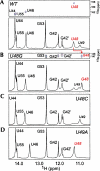
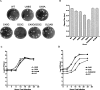
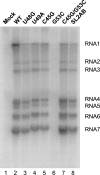
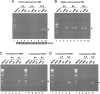
The 5' untranslated region (UTR) of the mouse hepatitis virus (MHV) genome contains cis-acting sequences necessary for transcription and replication. A consensus secondary structural model of the 5' 140 nucleotides of the 5' UTRs of nine coronaviruses (CoVs) derived from all three major CoV groups is presented and characterized by three major stem-loops, SL1, SL2, and SL4. NMR spectroscopy provides structural support for SL1 and SL2 in three group 2 CoVs, including MHV, BCoV, and HCoV-OC43. SL2 is conserved in all CoVs, typically containing a pentaloop (C47-U48-U49-G50-U51 in MHV) stacked on a 5 base-pair stem, with some sequences containing an additional U 3' to U51; SL2 therefore possesses sequence features consistent with a U-turn-like conformation. The imino protons of U48 in the wild-type RNA, and G48 in the U48G SL2 mutant RNA, are significantly protected from exchange with solvent, consistent with a hydrogen bonding interaction critical to the hairpin loop architecture. SL2 is required for MHV replication; MHV genomes containing point substitutions predicted to perturb the SL2 structure (U48C, U48A) were not viable, while those that maintain the structure (U48G and U49A) were viable. The U48C MHV mutant supports both positive- and negative-sense genome-sized RNA synthesis, but fails to direct the synthesis of positive- or negative-sense subgenomic RNAs. These data support the existence of the SL2 in our models, and further suggest a critical role in coronavirus replication.
Figures


References
-
- Cabello-Villegas, J., Tworowska, I., Nikonowicz, E.P. Metal ion stabilization of the U-turn of the A37 N6-dimethylallyl-modified anticodon stem–loop of Escherichia coli tRNAPhe. Biochemistry. 2004;43:55–66. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
