Amplified expression profiling of platelet transcriptome reveals changes in arginine metabolic pathways in patients with sickle cell disease
- PMID: 17353439
- PMCID: PMC2225987
- DOI: 10.1161/CIRCULATIONAHA.106.658641
Amplified expression profiling of platelet transcriptome reveals changes in arginine metabolic pathways in patients with sickle cell disease
Abstract
Background: In sickle cell disease, ischemia-reperfusion injury and intravascular hemolysis produce endothelial dysfunction and vasculopathy characterized by reduced nitric oxide and arginine bioavailability. Recent functional studies of platelets in patients with sickle cell disease reveal a basally activated state, which suggests that pathological platelet activation may contribute to sickle cell disease vasculopathy.
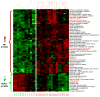
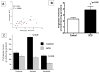
Methods and results: Studies were therefore undertaken to examine transcriptional signaling pathways in platelets that may be dysregulated in sickle cell disease. We demonstrate and validate in the present study the feasibility of comparative platelet transcriptome studies on clinical samples from single donors by the application of RNA amplification followed by microarray-based analysis of 54,000 probe sets. Data mining an existing microarray database, we identified 220 highly abundant genes in platelets and a subset of 72 relatively platelet-specific genes, defined by >10-fold increased expression compared with the median of other cell types in the database with amplified transcripts. The highly abundant platelet transcripts found in the present study included 82% or 70% of platelet-abundant genes identified in 2 previous gene expression studies on nonamplified mRNA from pooled or apheresis samples, respectively. On comparing the platelet gene expression profiles in 18 patients with sickle cell disease in steady state to those of 12 black control subjects, at a 3-fold cutoff and 5% false-discovery rate, we identified approximately 100 differentially expressed genes, including multiple genes involved in arginine metabolism and redox homeostasis. Further characterization of these pathways with real-time polymerase chain reaction and biochemical assays revealed increased arginase II expression and activity and decreased platelet polyamine levels.
Conclusions: The present studies suggest a potential pathogenic role for platelet arginase and altered arginine and polyamine metabolism in sickle cell disease and provide a novel framework for the study of disease-specific platelet biology.
Figures



References
-
- Bunn HF. Pathogenesis and treatment of sickle cell disease. N Engl J Med. 1997;337:762–769. - PubMed
-
- Reiter CD, Gladwin MT. An emerging role for nitric oxide in sickle cell disease vascular homeostasis and therapy. Curr Opin Hematol. 2003;10:99–107. - PubMed
-
- Castro O, Hoque M, Brown BD. Pulmonary hypertension in sickle cell disease: cardiac catheterization results and survival. Blood. 2003;101:1257–1261. - PubMed
-
- Gladwin MT, Sachdev V, Jison ML, Shizukuda Y, Plehn JF, Minter K, Brown B, Coles WA, Nichols JS, Ernst I, Hunter LA, Blackwelder WC, Schechter AN, Rodgers GP, Castro O, Ognibene FP. Pulmonary hypertension as a risk factor for death in patients with sickle cell disease. N Engl J Med. 2004;350:886–895. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

