Functioning and robustness of a bacterial circadian clock
- PMID: 17353932
- PMCID: PMC1847943
- DOI: 10.1038/msb4100128
Functioning and robustness of a bacterial circadian clock
Abstract
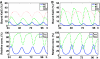
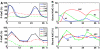
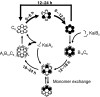
Cyanobacteria are the simplest known cellular systems that regulate their biological activities in daily cycles. For the cyanobacterium Synechococcus elongatus, it has been shown by in vitro and in vivo experiments that the basic circadian timing process is based on rhythmic phosphorylation of KaiC hexamers. Despite the excellent experimental work, a full systems level understanding of the in vitro clock is still lacking. In this work, we provide a mathematical approach to scan different hypothetical mechanisms for the primary circadian oscillator, starting from experimentally established molecular properties of the clock proteins. Although optimised for highest performance, only one of the in silico-generated reaction networks was able to reproduce the experimentally found high amplitude and robustness against perturbations. In this reaction network, a negative feedback synchronises the phosphorylation level of the individual hexamers and has indeed been realised in S. elongatus by KaiA sequestration as confirmed by experiments.
Figures




References
-
- Barkai N, Leibler S (1997) Robustness in simple biochemical networks. Nature 387: 913. - PubMed
-
- Elowitz MB, Levine AJ, Sigga ED, Swain PS (2002) Stochastic gene expression in a single cell. Science 297: 1183. - PubMed
-
- Hayashi F, Ito H, Fujita M, Iwase R, Uzumaki T, Ishiura M (2004) Stoichiometric interactions between cyanobacterial clock proteins KaiA and KaiC. Biochem Biophys Res Commun 316: 195. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

