Were inefficient mitochondrial haplogroups selected during migrations of modern humans? A test using modular kinetic analysis of coupling in mitochondria from cybrid cell lines
- PMID: 17355224
- PMCID: PMC1868799
- DOI: 10.1042/BJ20061609
Were inefficient mitochondrial haplogroups selected during migrations of modern humans? A test using modular kinetic analysis of coupling in mitochondria from cybrid cell lines
Abstract
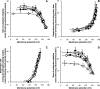
We introduce a general test of the bioenergetic importance of mtDNA (mitochondrial DNA) variants: modular kinetic analysis of oxidative phosphorylation in mitochondria from cybrid cells with constant nuclear DNA but different mtDNA. We have applied this test to the hypothesis [Ruiz-Pesini, Mishmar, Brandon, Procaccio and Wallace (2004) Science 303, 223-226] that particular mtDNA haplogroups (specific combinations of polymorphisms) that cause lowered coupling efficiency, leading to generation of less ATP and more heat, were positively selected during radiations of modern humans into colder climates. Contrary to the predictions of this hypothesis, mitochondria from Arctic haplogroups had similar or even greater coupling efficiency than mitochondria from tropical haplogroups.
Figures



Comment in
-
Testing the adaptive selection of human mtDNA haplogroups: an experimental bioenergetics approach.Biochem J. 2007 Jun 1;404(2):e3-5. doi: 10.1042/BJ20070524. Biochem J. 2007. PMID: 17488234 Free PMC article.
References
-
- Dimauro S., Davidzon G. Mitochondrial DNA and disease. Ann. Med. 2005;37:222–232. - PubMed
-
- Montiel-Sosa F., Ruiz-Pesini E., Enriquez J. A., Marcuello A., Diez-Sanchez C., Montoya J., Wallace D. C., Lopez-Perez M. J. Differences of sperm motility in mitochondrial DNA haplogroup U sublineages. Gene. 2006;368:21–27. - PubMed
-
- Baudouin S. V., Saunders D., Tiangyou W., Elson J. L., Poynter J., Pyle A., Keers S., Turnbull D. M., Howell N., Chinnery P. F. Mitochondrial DNA and survival after sepsis: a prospective study. Lancet. 2005;366:2118–2121. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

